Chapter 4 Example with real data
Let us use our prior knowledge on general linear models to explore the relationship between variables within the Oribatid mite data set.
Let us begin by loading this data set into R:
# Use setwd() to set your working directory
mites <- read.csv('data/mites.csv',
stringsAsFactors = TRUE)The dataset that you just loaded is a subset from the classic Oribatid mites (Acari,Oribatei), which has been used in numerous texts (e.g. Borcard,
Gillet & Legendre, Numerical Ecology with R), and which is available in the vegan library.
The Oribatid mite dataset has 70 observations with moss and mite samples collected at the Station de Biologie from the Université de Montréal. ], within the municipality of Saint-Hippolyte, Québec (Canada). Each sample includes 5 variables of environmental measurements and abundance for Galumna sp. for each site.
We can peek into the structure and the first six rows of the dataset using the head() and str() functions:
head(mites)## Galumna pa totalabund prop SubsDens WatrCont Substrate Shrub Topo
## 1 8 1 140 0.057142857 39.18 350.15 Sphagn1 Few Hummock
## 2 3 1 268 0.011194030 54.99 434.81 Litter Few Hummock
## 3 1 1 186 0.005376344 46.07 371.72 Interface Few Hummock
## 4 1 1 286 0.003496503 48.19 360.50 Sphagn1 Few Hummock
## 5 2 1 199 0.010050251 23.55 204.13 Sphagn1 Few Hummock
## 6 1 1 209 0.004784689 57.32 311.55 Sphagn1 Few Hummockstr(mites)## 'data.frame': 70 obs. of 9 variables:
## $ Galumna : int 8 3 1 1 2 1 1 1 2 5 ...
## $ pa : int 1 1 1 1 1 1 1 1 1 1 ...
## $ totalabund: int 140 268 186 286 199 209 162 126 123 166 ...
## $ prop : num 0.05714 0.01119 0.00538 0.0035 0.01005 ...
## $ SubsDens : num 39.2 55 46.1 48.2 23.6 ...
## $ WatrCont : num 350 435 372 360 204 ...
## $ Substrate : Factor w/ 7 levels "Barepeat","Interface",..: 4 3 2 4 4 4 4 2 3 4 ...
## $ Shrub : Factor w/ 3 levels "Few","Many","None": 1 1 1 1 1 1 1 2 2 2 ...
## $ Topo : Factor w/ 2 levels "Blanket","Hummock": 2 2 2 2 2 2 2 1 1 2 ...Our first glance into the dataset already allows us to separate potential response variables from potential predictors:
Response variables:
- Occurrence:
pa - Abundance:
Galumna - Relative Frequency or Proportion:
prop
Predictive variables:
- Substract Density:
SubsDens - Water Content:
WatrCont - Substrate:
Substrate - Shrubs Nearby:
Shrub - Topography:
Topo
We can also already elaborate an initial question: Could the abundance, occurrence or proportion of Galumna sp. be predicted by environmental features?
To answer this question, we can think of a variety of functions:
\(\text{Abundance} = f(\text{Water content}, \epsilon)\)
\(\text{Proportion} = f(\text{Water content}, \epsilon)\)
\(\text{Occurrence} = f(\text{Substrate}, \epsilon)\)
\(\text{Abundance} = f(\text{Topography}, \epsilon)\)
\(\text{Occurrence} = f(\text{Shrubs Nearby}, \epsilon)\)
\(\text{Relative Frequency} = f(\text{Topography}, \epsilon)\)
\(\text{Occurrence} = f(\text{Substract Density}, \epsilon)\)
\(\text{Abundance} = f(\text{Substrate}, \epsilon)\)
Can we see a relationship between Galumna and any of the five environmental variables?
Let us attempt to be more specific and ask whether he composition of Galumna’s communities (abundance, occurrence and relative frequency) vary as a function of water content?.
We can begin by representing all three response variables against the predictor:
plot(Galumna ~ WatrCont,
data = mites,
xlab = 'Water content',
ylab = 'Abundance')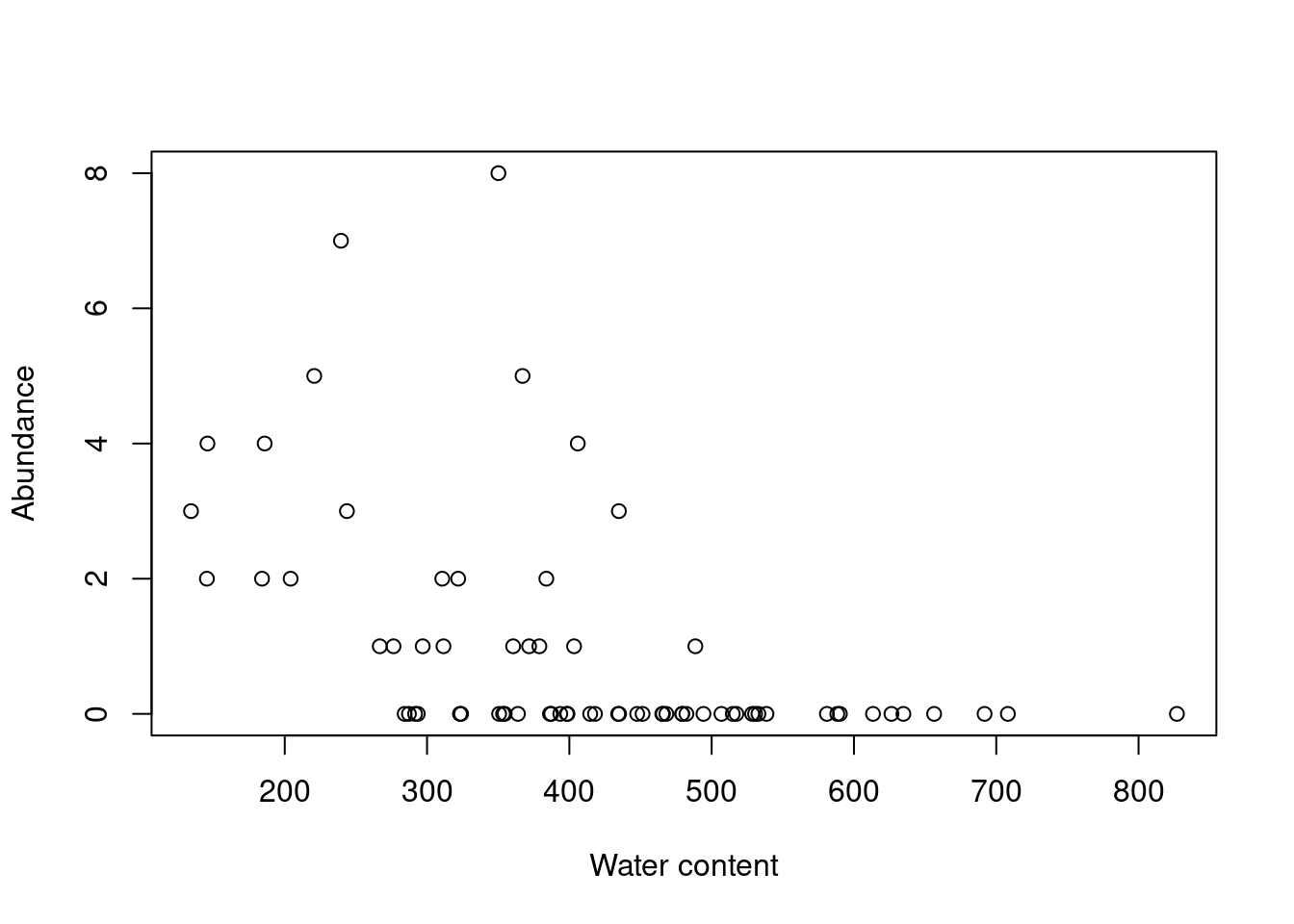
boxplot(WatrCont ~ pa,
data = mites,
xlab='Presence/Absence',
ylab = 'Water content')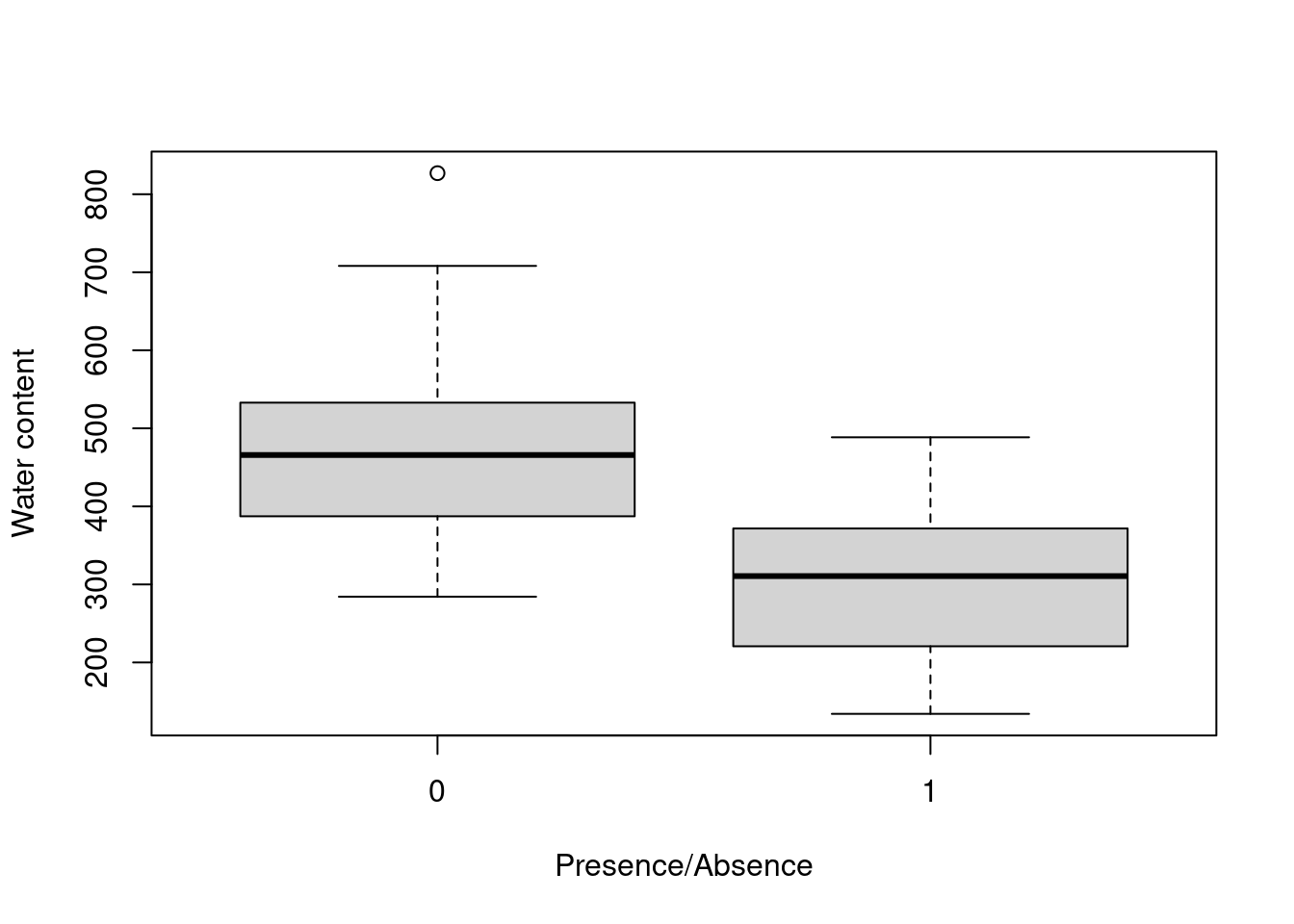
plot(prop ~ WatrCont,
data = mites,
xlab = 'Water content',
ylab = 'Proportion')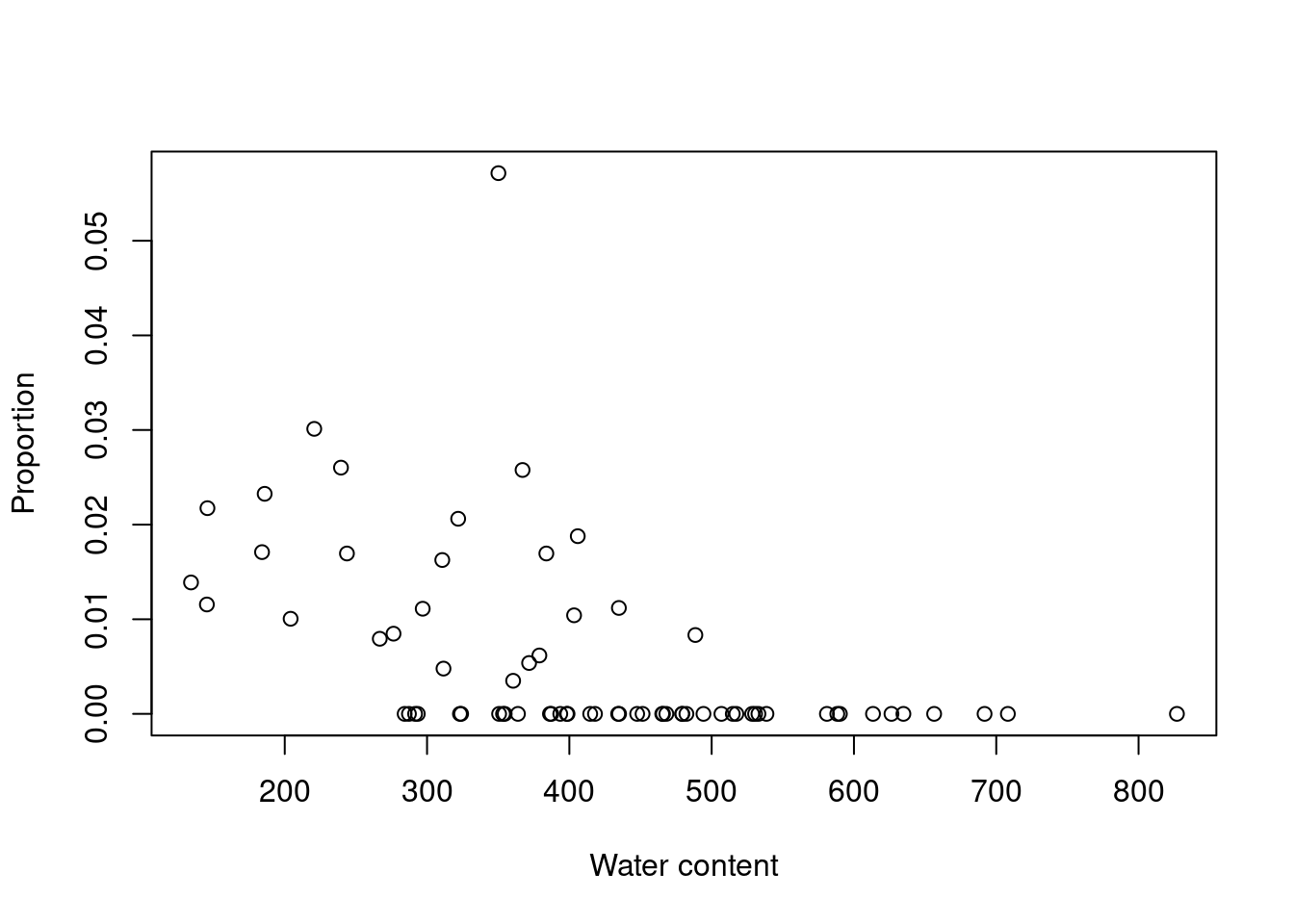
Indeed, Galumna seems to vary negatively as a function of WatrCont, i.e. Galumna sp. seems to prefer dryer sites.
We can go step further and fit general linear models to test whether Galumna, pa, or prop vary as a function of WatrCont using the lm() function:
# Fit the models
##
# Abundance model
lm.abund <- lm(Galumna ~ WatrCont, data = mites)
##
# Presence-absence model
lm.pa <- lm(pa ~ WatrCont, data = mites)
##
# Proportion model
lm.prop <- lm(prop ~ WatrCont, data = mites)Then, we can check the model output to verify whether these relationships are statistically significant:
# Check the model output with the summary() function
summary(lm.abund)##
## Call:
## lm(formula = Galumna ~ WatrCont, data = mites)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.7210 -0.8236 -0.3270 0.3910 6.6772
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 3.439349 0.555825 6.188 3.98e-08 ***
## WatrCont -0.006045 0.001280 -4.723 1.21e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.514 on 68 degrees of freedom
## Multiple R-squared: 0.247, Adjusted R-squared: 0.2359
## F-statistic: 22.31 on 1 and 68 DF, p-value: 1.206e-05summary(lm.pa)##
## Call:
## lm(formula = pa ~ WatrCont, data = mites)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.61320 -0.30889 -0.05498 0.30247 0.80073
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.1892142 0.1431306 8.309 6.03e-12 ***
## WatrCont -0.0020263 0.0003296 -6.148 4.68e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.3897 on 68 degrees of freedom
## Multiple R-squared: 0.3573, Adjusted R-squared: 0.3478
## F-statistic: 37.8 on 1 and 68 DF, p-value: 4.677e-08summary(lm.prop)##
## Call:
## lm(formula = prop ~ WatrCont, data = mites)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.010208 -0.004927 -0.002056 0.003240 0.049252
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 2.020e-02 3.294e-03 6.133 4.98e-08 ***
## WatrCont -3.516e-05 7.586e-06 -4.635 1.67e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.008971 on 68 degrees of freedom
## Multiple R-squared: 0.2401, Adjusted R-squared: 0.2289
## F-statistic: 21.49 on 1 and 68 DF, p-value: 1.665e-05# Extracting the Pr(>|t|)
summary(lm.abund)$coefficients[, 4]## (Intercept) WatrCont
## 3.981563e-08 1.206117e-05summary(lm.pa)$coefficients[, 4]## (Intercept) WatrCont
## 6.030252e-12 4.676755e-08summary(lm.prop)$coefficients[, 4]## (Intercept) WatrCont
## 4.977432e-08 1.665437e-05For now, yes, there is a strong and significant relationship for all 3 response variables!
But, wait a minute… We are forgetting something important here!
What about the assumptions of linear models?