class: center, middle, inverse, title-slide .title[ # Workshop 6: Generalized linear models ] .subtitle[ ## QCBS R Workshop Series ] .author[ ### Québec Centre for Biodiversity Science ] --- class: inverse, center, middle # About this workshop [](https://r.qcbs.ca/workshop06/pres-en/workshop06-pres-en.html) [](https://r.qcbs.ca/workshop06/book-en/workshop06-script-en.R) [](https://r.qcbs.ca/workshop06/book-en/index.html) [](https://r.qcbs.ca/workshops/r-workshop-06/) [](https://github.com/QCBSRworkshops/workshop06) --- <p style="font-size:75%"> .center[ **QCBS members who contributed to this workshop** by modifying and improving its content as part of the <br> *Le*arning *a*nd *D*evelopment *A*ward ] .pull-left[ .right[ **2022** - **2021** - **2020** [Laurie Maynard](https://github.com/LaurieMaynard) <br> [Pedro Henrique P. Braga](https://github.com/pedrohbraga) <br> [Katherine Hébert](https://github.com/katherinehebert) <br> [Alex Arkilanian](https://github.com/aarkilanian) <br> [Mathieu Vaillancourt](mathieu.vaillancourt.2@ulaval.ca) <br> [Esteban Góngora](https://github.com/estebangongora) ] ] .pull-right[ .left[ **2019** - **2018** - **2017** [Azenor Bideault](https://github.com/Azenor) <br> [Willian Vieira](https://github.com/willvieira) <br> [Pedro Henrique P. Braga](https://github.com/pedrohbraga) <br> [Marie Hélène Brice](https://github.com/mhBrice) <br> [Kevin Cazelles](https://github.com/KevCaz) **2016** - **2015** - **2014** [Cédric Frenette Dussault]() <br> [Thomas Lamy]() <br> [Zofia Taranu](https://github.com/zetaranu) <br> [Vincent Fugère](https://github.com/VFugere) ] ] </p> <br><br> .center[ __If you would like to contribute too__, visit [r.qcbs.ca/contributing](https://r.qcbs.ca/contributing/) <br> and don't hesitate to [get in touch](mailto:qcbs.csbq.r@gmail.com) with us! ] --- ## Learning objectives 1. Distinguish generalized linear models from general linear models (including many of their equations!). 2. Identify situations for when the use of generalized linear models is appropriate. 3. Test assumptions for generalized linear models. 4. Implement and execute generalized linear models in binary, proportion and count data. 5. Validate, interpret and visualise results of generalized linear models. <br> .center[.alert[Important] <br> This workshop will build up your knowledge on models alongside with some of their important equations. *Do not run to the mountains!* This will be achieved progressively and you will feel proud of yourself after! ] --- # Required material To follow this workshop, you are required to have downloaded and installed [RStudio](https://rstudio.com/products/rstudio/download/#download) and [R](https://cran.rstudio.com/). <br> .pull-left[ You must also use these packages: 1. [ggplot2](https://cran.r-project.org/package=ggplot2) 3. [MASS](https://cran.r-project.org/package=MASS) 4. [vcdExtra](https://cran.r-project.org/package=vcdExtra) 5. [bbmle](https://cran.r-project.org/package=bbmle) 6. [DescTools](https://cran.r-project.org/package=DescTools) ] .pull-right[ To install them from CRAN, run: ```r install.packages( c("ggplot2", 'MASS', 'vcdExtra', 'bbmle', 'DescTools') ) ``` ] <br> We will also use the following data sets: - <a href="https://r.qcbs.ca/workshop06/pres-en/data/mites.csv">mites.csv</a> - <a href="https://r.qcbs.ca/workshop06/pres-en/data/faramea.csv">faramea.csv</a> --- class: inverse, center, middle # Preface ### Reviewing linear models --- # Answering research questions through modelling Much of our research focuses on investigating how patterns we observe can be explained by predictive variables. We are often looking for a function `\(f\)` that can explain a response variable ( `\(Y\)` ) *in terms of* one ( `\(X_1\)` ) or many other predictors ( `\(X_2\)`, `\(X_3\)`, `\(...\)` , `\(X_n\)` ): `$$Y = f(X_1)$$` -- The combination of predictive variables we have sampled will *never* fully explain `\(Y\)`. Because of this, there is always *unpredictable disturbance* in our models, _i.e._ the error `\(\epsilon\)`. As such, the error is an irrevocable part of our function: `$$Y = f(X_1, \epsilon)$$` -- exclude: true In [Workshop 4](https://qcbsrworkshops.github.io/workshop04/pres-en/workshop04-pres-en.html#1), we have learned how to use **general linear models** as `\(f(\cdot)\)` to describe the relationship between variables. They were: .center[ .pull-left[ .pull-left[ `\(t\)`-test ] .pull-right[ ANOVA ] ] .pull-right[ .pull-left[ Linear regression ] .pull-right[ ANCOVA ] ] ] --- # Revisiting linear models In [Workshop 4](https://qcbsrworkshops.github.io/workshop04/pres-en/workshop04-pres-en.html#1), we have learned how to use **general linear models** as `\(f(\cdot)\)` to describe the relationship between variables. The general form of our function `\(Y = f(X_1)\)` as a linear function can be represented by: -- `$$Y_i = \beta_0 + \beta_1X_i + \varepsilon_i$$` where: `\(Y_i\)` is the predicted value of a response variable `\(\beta_0\)` is the *unknown coefficient* **intercept** `\(\beta_1\)` is the *unknown coefficient* **slope** `\(X_i\)` is the value for the explanatory variable `\(\varepsilon_i\)` is the model residual drawn from a normal distribution with a varying mean but a constant variance. ??? When explaining this slide, provide a few examples of Y and X variables (e.g. number of species, sampling effort, climatic gradient). X can also be called independent variable, explanatory variable, predictor variable; Y can also be called dependent variable, response variable, explained variable, predicted variable; ϵ can also be called random component, the error term, the disturbance. --- # Revisiting linear models and their assumptions We have also learned that **linear models** only produce unbiased estimators (_i.e._ are only reliable) if they follow certain assumptions. Most importantly: 1\. The population can be described by a linear relationship: `$$Y = \beta_0 + \beta_1X_i + \varepsilon_i$$` -- 2\. The error term `\(\varepsilon\)` has the same variance given any value of the explanatory variable (_i.e._ homoskedasticity), and the error terms are not correlated across observations (_i.e._ no autocorrelation): .pull-left[ `$$\mathbb{V}{\rm ar} (\epsilon_i | \mathbf{X} ) = \sigma^2_\epsilon,\ \forall i = 1,..,N$$` ] .pull-right[ `$$\mathbb{C}{\rm ov} (\epsilon_i, \epsilon_j) = 0,\ i \neq j$$` ] -- <br><br> 3\. And, the residuals are normal: `$$\boldsymbol{\varepsilon} | \mathbf{X} \sim \mathcal{N} \left( \mathbf{0}, \sigma^2_\epsilon \mathbf{I} \right)$$` ??? Note that the error and Y are always dependent, thus the Cov(Y, error) is different than zero. --- class: middle .small[ .center[**WHY SO MANY EQUATIONS?**] <br> .center[*Wait! Do not lose hope! They are necessary and we will explain why!*] ] --- ## What do we mean by: .pull-left[ 1\. The population can be described by a linear relationship: `$$Y = \beta_0 + \beta_1X_i + \varepsilon$$` 2\. Homoskedasticity and no autocorrelation: `$$\mathbb{V}{\rm ar} (\epsilon_i | \mathbf{X} ) = \sigma^2_\epsilon,\ \forall i = 1,..,N$$` `$$\mathbb{C}{\rm ov} (\epsilon_i, \epsilon_j) = 0,\ i \neq j$$` 3\. The residuals are normal: `$$\boldsymbol{\varepsilon} | \mathbf{X} \sim \mathcal{N} \left( \mathbf{0}, \sigma^2_\epsilon \mathbf{I} \right)$$` ] .pull-right[ The estimations of general linear models as in `\(\widehat{Y} = \widehat{\beta}_0 + \widehat{\beta}_1 X + \hat\varepsilon\)` assumes that data is generated following these assumptions. <br> <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-2-1.png" width="432" style="display: block; margin: auto;" /> ] ??? Describe the figure to the participants, showing the fitted line and the residuals, and explaining how variance does not increase with either variables. --- ## Representing the assumptions Let us simulate 250 observational units that satisfy the assumptions we have seen and have `\(\epsilon_i \sim \mathcal{N}(0, 2^2), i = 1,...,250\)`. .pull-left[ ```r nSamples <- 250 ID <- factor(c(seq(1:nSamples))) PredVar <- runif(nSamples, min = 0, max = 50) simNormData <- data.frame( ID = ID, PredVar = PredVar, RespVar = (2*PredVar + rnorm(nSamples, mean = 0, sd = 2) ) ) # We have learned how to use lm() lm.simNormData <- lm(RespVar ~ PredVar, data = simNormData) ``` ] -- .pull-right[ ```r layout(matrix(c(1,2,3,4),2,2)) plot(lm.simNormData) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-4-1.png" width="432" style="display: block; margin: auto;" /> ] ??? Residuals: residual = observed y – model-predicted y Outliers: observations with large residuals, i.e. the observed value for a point is very different from the one predicted by the regression model. Leverage points: A leverage point is defined as an observation that has a value of x that is far away from the mean of x. Influential observations: An influential observation is defined as an observation that changes the slope of the line. Thus, influential points have a large influence on the fit of the model. One method to find influential points is to compare the fit of the model with and without each observation. 1. Allows one to check the assumption of linearity and homoscedasticity; 2. QQ-plot allows the comparison of the residuals to "ideal" normal observations; it allows determining if two data sets come from populations with a common distribution. 3. Scale-location plot (square rooted standardized residual vs. predicted value) is useful for checking the assumption of homoscedasticity; 4. Cook's Distance, which is a measure of the influence of each observation on the regression coefficients. --- # An example using general linear models: real data Let us use our prior knowledge on general linear models to explore the relationship between variables within the *Oribatid mite dataset*. .pull-left3[ Let us begin by loading this dataset: .small[ ```r mites <- read.csv('data/mites.csv', stringsAsFactors = TRUE) ``` ] ] .pull-right3[ And, exploring it using: ```r str(mites) # also try head(mites) ``` ] <br> <br> <br> -- .small[ ``` # 'data.frame': 70 obs. of 9 variables: # $ Galumna : int 8 3 1 1 2 1 1 1 2 5 ... # $ pa : int 1 1 1 1 1 1 1 1 1 1 ... # $ totalabund: int 140 268 186 286 199 209 162 126 123 166 ... # $ prop : num 0.05714 0.01119 0.00538 0.0035 0.01005 ... # $ SubsDens : num 39.2 55 46.1 48.2 23.6 ... # $ WatrCont : num 350 435 372 360 204 ... # $ Substrate : Factor w/ 7 levels "Barepeat","Interface",..: 4 3 2 4 4 4 4 2 3 4 ... # $ Shrub : Factor w/ 3 levels "Few","Many","None": 1 1 1 1 1 1 1 2 2 2 ... # $ Topo : Factor w/ 2 levels "Blanket","Hummock": 2 2 2 2 2 2 2 1 1 2 ... ``` ] .small[ The Oribatid mite dataset has **70 observations with moss and mite samples**, and **5 environmental** measurements and abundance for *Galumna* sp. for each site. ] ??? It is from an original dataset [mites Oribatid (Acari,Oribatei)](http://adn.biol.umontreal.ca/~numericalecology/data/oribates.html) from the [Station de Biologie de l'Université de Montréal](https://goo.gl/maps/PxN1Q7KUPnUt92Eu5). ] --- # Which potential questions can we make? **Question**: *Could the abundance, occurrence or proportion of Galumna sp. be predicted by environmental features?* .pull-left[ Response variables: 1. Occurrence: `pa` 2. Abundance: `Galumna` 3. Proportion or Relative Frequency: `prop` ] .pull-right[ Predictive variables: 1. Substract Density: `SubsDens` 2. Water Content: `WatrCont` 3. Substrate: `Substrate` 4. Shrubs Nearby: `Shrub` 5. Topography: `Topo` ] -- <br> **To answer our question**, we can develop models! *So many options!* .pull-left[ `\(\text{Abundance} = f(\text{Water content}, \epsilon)\)` `\(\text{Proportion} = f(\text{Water content}, \epsilon)\)` `\(\text{Occurrence} = f(\text{Substrate}, \epsilon)\)` `\(\text{Abundance} = f(\text{Topography}, \epsilon)\)` ] .pull-right[ `\(\text{Occurrence} = f(\text{Shrubs Nearby}, \epsilon)\)` `\(\text{Relative Frequency} = f(\text{Topography}, \epsilon)\)` `\(\text{Occurrence} = f(\text{Substract Density}, \epsilon)\)` `\(\text{Abundance} = f(\text{Substrate}, \epsilon)\)` ] --- exclude: true # Where do we begin? .small[Can we see a relationship between *Galumna* and any of the five environmental variables?] .pull-left2[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-9-1.png" width="576" style="display: block; margin: auto;" /> ] .pull-right2[ <br><br><br><br><br> ```r plot(mites) ``` What about... `Galumna` `\(vs\)` `WatrCont`?! ] --- # Exploring relationships Does the composition of *Galumna*'s communities (abundance, occurrence and relative frequency) vary as a function of water content? .small[ ```r par(mfrow = c(1, 3), cex = 1.4) plot(Galumna ~ WatrCont, data = mites, xlab = 'Water content', ylab = 'Abundance') boxplot(WatrCont ~ pa, data = mites, xlab = 'Presence-absence', ylab = 'Water content', col = 'red') plot(prop ~ WatrCont, data = mites, xlab = 'Water content', ylab = 'Relative frequency') ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-11-1.png" width="864" style="display: block; margin: auto;" /> ] ??? --- # Using linear models with `lm()` Let us use (general) linear models to test whether `Galumna`, `pa`, and/or `prop` vary as a function of `WatrCont` using the `lm()` function: -- .small[ .pull-left3[ ```r lm.abund <- lm(Galumna ~ WatrCont, data = mites) ## summary(lm.abund) lm.pa <- lm(pa ~ WatrCont, data = mites) ## summary(lm.pa) lm.prop <- lm(prop ~ WatrCont, data = mites) ## summary(lm.prop) ``` ] .pull-right3[ <br> `\(\text{Galumna} = f(\text{Water content}, \epsilon)\)` <br> `\(\text{Occurrence} = f(\text{Water content}, \epsilon)\)` <br> `\(\text{Proportion} = f(\text{Water content}, \epsilon)\)` ] ] -- .small[ .pull-left[ ```r # Extracting the Pr(>|t|) summary(lm.abund)$coefficients[, 4] # (Intercept) WatrCont # 3.981563e-08 1.206117e-05 summary(lm.pa)$coefficients[, 4] # (Intercept) WatrCont # 6.030252e-12 4.676755e-08 summary(lm.prop)$coefficients[, 4] # (Intercept) WatrCont # 4.977432e-08 1.665437e-05 ``` ] ] .center[ .pull-right[ <br> <br> All models are significant! <br> .alert[But...] **what about the assumptions?** ] ] --- # Using linear models: assumptions .alert[Wait a minute...] .small[ .pull-left[ ```r plot(Galumna ~ WatrCont, data = mites) abline(lm.abund) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-14-1.png" width="432" style="display: block; margin: auto;" /> ] ] .pull-right[ ```r par(mfrow = c(2, 2), cex = 1.4) plot(lm.abund) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-15-1.png" width="432" style="display: block; margin: auto;" /> ] ??? 1. Allows one to check the assumption of linearity and homoscedasticity; 2. QQ-plot allows the comparison of the residuals to "ideal" normal observations; it allows determining if two data sets come from populations with a common distribution. 3. Scale-location plot (square rooted standardized residual vs. predicted value) is useful for checking the assumption of homoscedasticity; 4. Cook's Distance, which is a measure of the influence of each observation on the regression coefficients. --- # Using linear models: assumptions They look very bad for `lm(prop ~ WatrCont)`: .pull-left[ ```r plot(prop ~ WatrCont, data = mites) abline(lm.prop) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-16-1.png" width="432" style="display: block; margin: auto;" /> ] .pull-right[ ```r par(mfrow = c(2, 2), cex = 1.4) plot(lm.prop) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-17-1.png" width="432" style="display: block; margin: auto;" /> ] --- # Using linear models: assumptions And, also bad for `lm(pa ~ WatrCont)`: .pull-left[ ```r plot(pa ~ WatrCont, data = mites) abline(lm.pa) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-18-1.png" width="432" style="display: block; margin: auto;" /> ] .pull-right[ ```r par(mfrow = c(2, 2), cex = 1.4) plot(lm.pa) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-19-1.png" width="432" style="display: block; margin: auto;" /> ] --- # Recalling linear models: assumptions .small[ .pull-left[ Remember our simple linear model? `$$Y_i = \beta_0 + \beta_1X_i + \varepsilon$$` It can also be written as this: `$$Y_i \sim N(\mu = \beta_0 + \beta_1 X_i +\varepsilon, \sigma^2)$$` with `\(N(\cdot)\)` meaning that `\(Y_i\)` is drawn from a **normal distribution** with parameters `\(\mu\)` (mean; which depends on `\(x_i\)`) and `\(\sigma\)` (variance; which has the same value for all `\(Y_i\)`). ] .pull-right[ Varying `\(\mu\)`, `\(\sigma = 5\)` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-20-1.png" width="216" style="display: block; margin: auto;" /> `\(\mu = 25\)`, varying `\(\sigma\)` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-21-1.png" width="216" style="display: block; margin: auto;" /> ] ] ??? On the right, we show graphs where mu varies or sigma varies. The top figure corresponds shows that sigma is constant across mean values of Y. This is closer to linear assumptions The bottom figure exacerbates a situation where sigma increases across the mean of y. --- # Model prediction Remember that we aim at estimating the *unknown coefficients* `\(\beta_0\)` and `\(\beta_1\)`, so that a line effectively predicting every value of `\(Y\)` as a function of `\(X\)` can be drawn! `$$Y_i \sim N(\mu = \beta_0 + \beta_1 X_i +\varepsilon, \sigma^2)$$` We can obtain the parameters `\(\mu\)` and `\(\sigma^2\)` for a normal distribution corresponding to our equation! To obtain the coefficients from our models, we can use the function `coef()`: .pull-left[ ```r coef(lm.abund) # (Intercept) WatrCont # 3.439348672 -0.006044788 ``` ] .pull-right[ ```r summary(lm.abund)$sigma # [1] 1.513531 ``` ] What are the _predicted_ parameters of the normal distribution used to model `\(Y\)` when water content is `\(300\)`? -- .center[ for `\(X_{i} = 300\)`, `\(\mu = 3.44 + (-0.006 \times 300) = 1.63\)` and `\(\sigma^2 = 1.51\)` ] _i.e._ randomly drawn `\(Y\)` values when water content is `\(300\)` should be on average `\(1.63\)` and have variance `\(1.51\)`. --- # Model prediction .pull-left[ `$$y_i \sim N(\mu = \beta_0 + \beta_1 X_i +\varepsilon, \sigma^2)$$` `$$\mu = 3.44 + (-0.006 \times 400) +\varepsilon= 1.02$$` `$$\sigma^2 = 1.51$$` ] .pull-right[ At `\(x = 300\)`, residuals should follow a normal distribution with `\(\mu = 1.63\)` and `\(\sigma^2 = 1.51\)`. At `\(x = 400\)`, we expect `\(\mu = 1.02\)` and `\(\sigma^2 = 1.51\)`, and there on. ] -- .pull-left[ .small[Our model thus expects observations to fall within the following shaded areas]: .center[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-22-1.png" width="324" style="display: block; margin: auto;" /> ] ] -- <br> .pull-right[ .center[**But, do they?**] ] --- # Model prediction .pull-left[ `$$y_i \sim N(\mu = \beta_0 + \beta_1 X_i +\varepsilon, \sigma^2)$$` `$$\mu = 3.44 + (-0.006 \times 400) +\varepsilon= 1.02$$` `$$\sigma^2 = 1.51$$` .small[Our model thus expects observations to fall within the following shaded areas]: .center[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-23-1.png" width="324" style="display: block; margin: auto;" /> ] ] .pull-right[ At `\(x = 300\)`, residuals should follow a normal distribution with `\(\mu = 1.63\)` and `\(\sigma^2 = 1.51\)`. At `\(x = 400\)`, we expect `\(\mu = 1.02\)` and `\(\sigma^2 = 1.51\)`, and there on. <br> .center[**But, do they?**] .center[*Hmmmmmmmm...*] ] --- # Model prediction .pull-left[ `$$y_i \sim N(\mu = \beta_0 + \beta_1 X_i +\varepsilon, \sigma^2)$$` `$$\mu = 3.44 + (-0.006 \times 400) +\varepsilon= 1.02$$` `$$\sigma^2 = 1.51$$` .small[Our model thus expects observations to fall within the following shaded areas]: .center[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-24-1.png" width="324" style="display: block; margin: auto;" /> ] ] .pull-right[ At `\(x = 300\)`, residuals should follow a normal distribution with `\(\mu = 1.63\)` and `\(\sigma^2 = 1.51\)`. At `\(x = 400\)`, we expect `\(\mu = 1.02\)` and `\(\sigma^2 = 1.51\)`, and there on. <br> .center[**But, do they?**] .center[*Hmmmmmmmm...*] <br> **Which of the following are correct?** 1. The residuals seem to violate linear assumptions; 2. `\(\sigma^2\)` (variance) changes across the values of `\(x\)`; 3. The observations do not fall across the expected values. ] ??? You can perform a multiple choice poll with participants. Options 1, 2 and 3 are problems in this model, i.e. all are correct. --- ## What do we do now? Transform? Very often, data will not "behave" and will violate the parametric assumptions, showing evidence for **non-normality** and/or **heteroskedasticity**. We can **transform** our data using logarithmic, square-root, and cosine (and other) transformations. -- .pull-left[ But, transformations not always work and come with a few drawbacks: **1.** They change the response variable (!), making interpretation challenging; **2.** They may not simulateneously improve linearity and homogeneity of variance; **3.** The boundaries of the sample space change. ] -- .pull-right[ For instance, our simple linear model: `$$Y_i = \beta_0 + \beta_1X_i + \varepsilon$$` looks like this under a log-transform: $$E(\log{Y_i}) = \beta_0 + \beta_1X_i $$ *It seems undesirable to interpret that for every `\(300\)` units increase in water content, Galumna abundance takes the form of `\(\log(1.63).\)`* ] --- # Normal distributions are not the only ones! Statisticians have defined a [multitude of probability distributions](https://www.causascientia.org/math_stat/Dists/Compendium.pdf) to describe different types of data. A distribution provides the probability of observing each possible outcome of an experiment or survey (_e.g._ `\(abund = 8\)` *Galumna* individuals). -- Distributions can be **discrete** (only include integers), **continuous** (including fractions) and be limited to a given range or domain (_e.g._, `\(0\)` to `\(1\)`, `\(0\)` to `\(+\infty\)`, `\(-\infty\)` to `\(+\infty\)`). <br> All distributions can be broken down into **parameters** that dictate their shapes (_e.g._, `\(\mu\)` and `\(\sigma^2\)` for the Normal). --- # Biological data & distributions: Poisson *Galumna* `abund` follows a discrete distribution (_i.e._ it only takes integer values). Abundance data can often be described through the “Poisson” distribution: - **Poisson** is a discrete distribution with a single parameter, `\(\lambda\)` (lambda), which defines both the mean and the variance of the distribution: <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-25-1.png" width="1080" style="display: block; margin: auto;" /> --- # Biological data & distributions: Poisson .pull-left[ *Galumna* seems to follow a Poisson distribution with a low value of `\(\lambda\)`: ```r hist(mites$Galumna) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-26-1.png" width="432" style="display: block; margin: auto;" /> ] .pull-right[ Would a model incorporating the Poisson distribution work better in the `\(\text{Galumna} = f(\text{Water content})\)` case? <br> <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-27-1.png" width="432" style="display: block; margin: auto;" /> ] ??? Again, note how we can interpret this where the values are expected to follow around the orange lines (before it was shaded because the values could be continuous; here they are discrete). --- # Biological data & distributions Presence-absence data takes another form: - Only `\(0\)` and `\(1\)`; - Poisson distribution would not be appropriate to model this variable. ```r hist(mites$pa) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-28-1.png" width="432" style="display: block; margin: auto;" /> --- # Biological data & distributions: Bernoulli **“Bernoulli” distribution**: - Only two possible outcomes in its range: success (`1`) or failure (`0`); - One parameter, `\(p\)`, the probability of success. <br> <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-29-1.png" width="864" style="display: block; margin: auto;" /> We can use the Bernouilli distribution to calculate the frequency probability for the presence of _Galumna_ (`1`) in relation to its absence (`0`). ??? A Bernoulli event is one for which the probability the event occurs is p and the probability the event does not occur is 1-p; i.e., the event is has two possible outcomes (usually viewed as success or failure) occurring with probability p and 1-p, respectively. A Bernoulli distribution is the pair of probabilities of a Bernoulli event, which is too simple to be interesting. However, it is implicitly used in “yes, no” decision processes where the choice occurs with the same probability from trial to trial (e.g., the customer chooses to go down aisle 1 with probability p). --- # Biological data & distributions: Binomial **Binomial distribution**: When there are multiple trials (each with a success/failure), the Bernoulli distribution expands into the binomial distribution. - Additional parameter, `\(n\)`, for number of trials; - Predicts the probability of observing a given proportion of successes, `\(p\)`, out of a known total number of trials, `\(n\)`. <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-30-1.png" width="1080" style="display: block; margin: auto;" /> --- # Biological data & distributions: Binomial **Binomial distribution**: used to model data where the number of successes are integers and where the number of trials, `\(n\)`, is known. **Main difference with Poisson distribution**: the binomial has an upper limit to its range, corresponding to `\(n\)`. Consequently, it is right-skewed at low `\(p\)` values but left-skewed at high `\(p\)` values <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-31-1.png" width="720" style="display: block; margin: auto;" /> ??? Now, while the Bernoulli distribution represents the success or failure of a single Bernoulli trial. The Binomial Distribution represents the number of successes and failures in n independent Bernoulli trials for some given value of n. A Bernoulli random variable has two possible outcomes: 0 or 1. A binomial distribution is the sum of independent and identically distributed Bernoulli random variables. --- class: middle, center *Getting back to our problem... what do we do if we have other distributions?* <br> .center[We can try using models that are able to incorporate other types of distributions: the **generalized linear models**!] <br> In `R`, this means that we will move away from the (general) linear models function `lm()` and begin using the `glm()` function! --- # Generalized Linear Models A **generalized linear model** is made of a **linear predictor**: `$$\eta_i = \underbrace{g(\mu)}_{Link~~function} = \underbrace{\beta_0 + \beta_1X_1~+~...~+~\beta_pX_p}_{Linear~component}$$` and, 1. a **link** function `\(g(\mu_i)\)` that ***transforms the expected values*** `\(E(Y_i)\)`, and describes how the mean of the process `\(Y\)`, `\(E(Y_i) = \mu_i\)`, depends on the linear predictor `$$g(\mu_i) = ηi$$` 2\. a **variance** function that describes how the variance of the process `\(Y\)`, `\(\text{var}(Y_i)\)`, depends on the mean `$$\text{var}(Y_i) = \phi V(\mu)$$` where the **dispersion parameter** `\(\phi\)` (_phi_) is a constant. ??? The link function relates the expected value of the response to the linear predictors in the model, so that the relationship between the predictors and the response can be modeled with linear regression. How is that different from transforming the variable? We often log-transform data because the values of y do not appear to be normally distributed around their average. When we transform the data in general linear models, we are no longer claiming that y is normally distributed around a mean, given the x values; we are now claiming that our new outcome variable, ln(yi) (if we used log), is normally distributed. In the case of the glm Poisson model, however, the link function does not change the distribution of the actual observations in some way to make them something other than Poisson distributed. Instead, the link function defines the relationship of the x variables directly to the mean of the Poisson distributed y. The individual observations then vary around this expected value accordingly. The mean of the log is not the log of the mean: one may not take the exponent of the mean of ln(y) to get the mean of y. On the other hand, the link function allows you to do that! --- # General linear models as a GLM So, for our **general linear model** with `\(\epsilon ∼ N(0, σ^2)\)` and `\(Y_i \sim{} N(\mu_i, \sigma^2)\)`, we have the same linear predictor: `$$\underbrace{g(\mu)}_{Link~~function} = \underbrace{\beta_0 + \beta_1X_1~+~...~+~\beta_pX_p}_{Linear~component}$$` the link function: `$$g(\mu_i) = \mu_i$$` and, the variance function: `$$V(\mu_i) = 1$$` <br> .center[*Wow! This is equivalent to the normal linear model that we have learned before!*] --- # `glm()` In `R`, we can fit generalized linear models using the `glm()` function, which is similar to the `lm()` function: .pull-left2[ ```r glm(formula, family = gaussian(link = "identity"), data, ...) ``` ] .pull-right2[ ```r lm(formula, data, ...) # ``` ] <br> with the `family` argument taking the names of the **link function** (inside `link`) and, the **variance function**. This approach is extendable to other distribution families! ??? The link function for a general linear model is the identity link (see next slide) -- | Distribution of `\(Y\)` | Link function name | Link function | Model | `R` | |---------------------|--------------------|---------------|------------|-----| | Normal | Identity | `\(g(\mu) = \mu\)` | `\(\mu = \mathbf{X} \boldsymbol{\beta}\)` | `gaussian(link="identity")` | | Binomial | Logit | `\(g(\mu) = \log\left(\dfrac{\mu}{1-\mu}\right)\)` | `\(\log\left(\dfrac{\mu}{1-\mu}\right) = \mathbf{X} \boldsymbol{\beta}\)` | `binomial(link="logit")`| | Poisson | Log | `\(g(\mu) = \log(\mu)\)` | `\(\log(\mu) = \mathbf{X} \boldsymbol{\beta}\)` | `poisson(link="log")`| | Exponential | Negative Inverse | `\(g(\mu) = -\mu^{-1}\)` | `\(-\mu^{-1} = \mathbf{X} \boldsymbol{\beta}\)` | `Gamma(link="inverse")` | ??? How is that different from transforming the variable? We often log-transform data because the values of y do not appear to be normally distributed around their average. When we transform the data in general linear models, we are no longer claiming that y is normally distributed around a mean, given the x values; we are now claiming that our new outcome variable, ln(yi) (if we used log), is normally distributed. In the case of the glm Poisson model, however, the link function does not change the distribution of the actual observations in some way to make them something other than Poisson distributed. Instead, the link function defines the relationship of the x variables directly to the mean of the Poisson distributed y. The individual observations then vary around this expected value accordingly. The mean of the log is not the log of the mean: one may not take the exponent of the mean of ln(y) to get the mean of y. On the other hand, the link function allows you to do that! --- class: inverse, center, middle # GLM with binary data ## Generalized linear models --- # Binary variables A common response variable in ecological datasets is the **binary variable**. We observe a phenomenon `\(Y\)` or its “absence”: - Presence or absence of a species; - Presence or absence of a disease; - Success or failure to observe a certain behaviour; - Survival or death of organisms. <br> As usual, we are interested in questions such as _how species occurrences vary in function of the environment_? `$$\text{Occurrences} = f(\text{Environment})$$` --- # Binary variables Under a linear model, expected values can be out of the `[0, 1]` range with `lm()`: <br> ```r lm(Pres ~ ExpVar) ``` <br> .pull-left[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-35-1.png" width="360" style="display: block; margin: auto;" /> ] .pull-right[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-36-1.png" width="504" style="display: block; margin: auto;" /> ] --- # GLM with binomial data: logit link A **generalized linear model** is made of a **linear predictor**: `$$\underbrace{g(\mu_i)}_{Link~~function} = \underbrace{\beta_0 + \beta_1X_1~+~...~+~\beta_pX_p}_{Linear~component}$$` Consider that `\(Y_i ∼ B(n_i, p_i)\)`, and that we want to model the proportions of `\(Y_i^{}/n_i\)`. As such: .center[ `\(E(Y_i^{}/n_i) = p_i\)` and `\(\text{var}(Y_i^{}/n_i) = \frac{1}{n_i}p_i(1-p_i)\)` , so that `\(V(\mu_i) = \mu_i(1-\mu_i)\)`.] -- .pull-left[ We first move the probabilities `\(\mu_i\)` to the *odds*: `$$\text{odds}_i = \frac{\mu_i}{1-\mu_i}$$` The odds puts our expected values on a `0` to `+Inf` scale. ] -- .pull-right[ We then take logarithms, calculating the *logit* or log-odds: `$$\eta_i = \text{logit}(\mu_i) = \log(\frac{\mu_i}{1-\mu_i})$$` with `\(\mu\)` being the expected values (probability that `\(Y = 1\)` ), and with the expected values now ranging from `-Inf` to `+Inf`. ] ??? We can understand that as if event A has probability p of occurring, then the odds of event A occurring is the ratio of the probability that A occurs to the probability that A does not occur: p/(1−p). For example, if the probability that I will fail my courses is 0.6, the odds that I will fail my courses is 0.6/(1 − 0.6) = 1.5. This means that the probability of observing a failure in my courses is 1.5 times greater than the probability of not observing it (that is, 1.5 × 0.4 = 0.6). --- # GLM for binary data: logit link A reminder about our `glm()` function: ```r glm(formula, family = ???, data, ...) ``` -- | Distribution of `\(Y\)` | Link function name | Link function | Model | `R` | |---------------------|--------------------|---------------|------------|-----| | Normal | Identity | `\(g(\mu) = \mu\)` | `\(\mu = \mathbf{X} \boldsymbol{\beta}\)` | `gaussian(link="identity")` | | Binomial | Logit | `\(g(\mu) = \log\left(\dfrac{\mu}{1-\mu}\right)\)` | `\(\log\left(\dfrac{\mu}{1-\mu}\right) = \mathbf{X} \boldsymbol{\beta}\)` | `binomial(link="logit")`| | Poisson | Log | `\(g(\mu) = \log(\mu)\)` | `\(-\mu^{-1} = \mathbf{X} \boldsymbol{\beta}\)` | `poisson(link="log")`| | Exponential | Negative Inverse | `\(g(\mu) = -\mu^{-1}\)` | `\(\log(\mu) = \mathbf{X} \boldsymbol{\beta}\)` | `Gamma(link="inverse")` | -- ```r glm(formula, family = binomial(link = "logit"), # this is also known as logistic data, ...) ``` --- # Exercise 1 - Our first generalized linear model, together. Build a logistic regression model using the `mites` data ```r # setwd('...') mites <- read.csv("data/mites.csv", header = TRUE) str(mites) # 'data.frame': 70 obs. of 9 variables: # $ Galumna : int 8 3 1 1 2 1 1 1 2 5 ... # $ pa : int 1 1 1 1 1 1 1 1 1 1 ... # $ totalabund: int 140 268 186 286 199 209 162 126 123 166 ... # $ prop : num 0.05714 0.01119 0.00538 0.0035 0.01005 ... # $ SubsDens : num 39.2 55 46.1 48.2 23.6 ... # $ WatrCont : num 350 435 372 360 204 ... # $ Substrate : chr "Sphagn1" "Litter" "Interface" "Sphagn1" ... # $ Shrub : chr "Few" "Few" "Few" "Few" ... # $ Topo : chr "Hummock" "Hummock" "Hummock" "Hummock" ... ``` ??? A reminder about the dataset: 70 sites (rows) x 35 morphospecies (columns). The sampling units are substrate cores 5 cm in diameter and 10 cm in depth, hereafter called "cores". SubsDens: substratum density in g.L-1 of dry uncompressed matter (quantitative variable) WatrCont: water content of the substratum, in percent of volume (quantitative variable) Borcard, D., P. Legendre and P. Drapeau. 1992. Partialling out the spatial component of ecological variation. Ecology 73: 1045-1055. Borcard, D. and P. Legendre. 1994. Environmental control and spatial structure in ecological communities: an example using Oribatid mites (Acari, Oribatei). Environmental and Ecological Statistics 1: 37-53. --- # Exercise 1 - Our first generalized linear model, together. Let us build a model of the presence of *Galumna* sp. as a function of water content and topography. ```r logit.reg <- glm(pa ~ WatrCont + Topo, data = mites, family = binomial(link = "logit")) ``` ```r summary(logit.reg) ``` ??? The presenter must highlight the `family` argument, explaining what it is. Also, refer to ?family. --- # Exercise 1 .small[ ```r summary(logit.reg) # # Call: # glm(formula = pa ~ WatrCont + Topo, family = binomial(link = "logit"), # data = mites) # # Deviance Residuals: # Min 1Q Median 3Q Max # -2.0387 -0.5589 -0.1594 0.4112 2.0252 # # Coefficients: # Estimate Std. Error z value Pr(>|z|) # (Intercept) 4.464402 1.670622 2.672 0.007533 ** # WatrCont -0.015813 0.004535 -3.487 0.000489 *** # TopoHummock 2.090757 0.735348 2.843 0.004466 ** # --- # Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 # # (Dispersion parameter for binomial family taken to be 1) # # Null deviance: 91.246 on 69 degrees of freedom # Residual deviance: 48.762 on 67 degrees of freedom # AIC: 54.762 # # Number of Fisher Scoring iterations: 6 ``` ] ??? When explaining this summary show what the blocks are and recall that its structure resembles the one from `summary.lm()`, but that there are some special differences (e.g. dispersion parameter). Deviance, coefficients and dispersion will be covered in the following slides. --- # Challenge 1 ![:cube]() Using the `bacteria` data set, model the presence of *H. influenzae* as a function of treatment and week of test. Start with a full model and reduce it to the most parsimonious model. ```r library(MASS) data(bacteria); str(bacteria) # 'data.frame': 220 obs. of 6 variables: # $ y : Factor w/ 2 levels "n","y": 2 2 2 2 2 2 1 2 2 2 ... # $ ap : Factor w/ 2 levels "a","p": 2 2 2 2 1 1 1 1 1 1 ... # $ hilo: Factor w/ 2 levels "hi","lo": 1 1 1 1 1 1 1 1 2 2 ... # $ week: int 0 2 4 11 0 2 6 11 0 2 ... # $ ID : Factor w/ 50 levels "X01","X02","X03",..: 1 1 1 1 2 2 2 2 3 3 ... # $ trt : Factor w/ 3 levels "placebo","drug",..: 1 1 1 1 3 3 3 3 2 2 ... ``` *Tests of the presence of the bacteria H. influenzae in children with otitis media in the Northern Territory of Australia.* - `y` is the presence or absence: a factor with levels `n` and `y`; - `week` is the week of test; - `trt` is the treatment ` placebo`, `drug` and `drug+`, a re-coding of `ap` and `hilo`. ??? Dr A. Leach tested the effects of a drug on 50 children with a history of otitis media in the Northern Territory of Australia. The children were randomized to the drug or a placebo. The presence of _H. influenzae was_ checked at weeks 0, 2, 4, 6 and 11: 30 of the checks were missing and are not included in this data frame. You can make this as a breakout room activity. You can assign groups of 3 - 4 people into breakout rooms and give them 5 minutes to collaboratively write the code. ] --- # Challenge 1 - Solution ![:cube]() ```r model.bact1 <- glm(y ~ trt * week, data = bacteria, family = binomial) ``` ```r model.bact2 <- glm(y ~ trt + week, data = bacteria, family = binomial) ``` ```r model.bact3 <- glm(y ~ week, data = bacteria, family = binomial) ``` ```r anova(model.bact1, model.bact2, model.bact3, test = "LRT") # Analysis of Deviance Table # # Model 1: y ~ trt * week # Model 2: y ~ trt + week # Model 3: y ~ week # Resid. Df Resid. Dev Df Deviance Pr(>Chi) # 1 214 203.12 # 2 216 203.81 -2 -0.6854 0.70984 # 3 218 210.91 -2 -7.1026 0.02869 * # --- # Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 plot(model.bact1) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-47-1.png" width="432" style="display: block; margin: auto;" /><img src="workshop06-pres-en_files/figure-html/unnamed-chunk-47-2.png" width="432" style="display: block; margin: auto;" /><img src="workshop06-pres-en_files/figure-html/unnamed-chunk-47-3.png" width="432" style="display: block; margin: auto;" /><img src="workshop06-pres-en_files/figure-html/unnamed-chunk-47-4.png" width="432" style="display: block; margin: auto;" /> ??? LRT performs a likelihood ratio (LR) test between the models. It is obtained as in the example below. # Improve this in the future vals <- (sum(residuals(base)^2) - sum(residuals(full)^2))/sum(residuals(full)^2) * full$df.residual pchisq(vals, df.diff, lower.tail = FALSE) --- # Interpreting the `glm()` coefficients Let's go back to the summary of our `logit.reg` model to see the coefficients: ```r logit.reg <- glm(pa ~ WatrCont + Topo, data=mites, family = binomial(link = "logit")) ``` ```r summary(logit.reg)$coefficients # Estimate Std. Error z value Pr(>|z|) # (Intercept) 4.46440199 1.670622482 2.672299 0.0075333598 # WatrCont -0.01581255 0.004535069 -3.486728 0.0004889684 # TopoHummock 2.09075654 0.735348234 2.843220 0.0044660283 ``` The output indicates that both water content and topography are significant drivers of the presence-absence variable. <br> .center[.comment[But, how do we interpret the slope coefficients in `glm()`?]] --- # Interpreting the `glm()` coefficients The direct interpretation of the coefficients in the logit model is tenuous because of the link function. If the link is *identity*, it is much easier to interpret. Let us assume that we have a binary outcome `\(y\)` and two covariates `\(x_1\)` and `\(x_2\)` (and a constant). The probability of a successful outcome ( `\(y = 1\)` ) is given by: `$$Pr(y_i = 1) = p = g^{-1}(\beta_0 + x_{1i}\beta_1 + x_{2i}\beta_2)$$` where `\(g^{-1}()\)` is the **inverse link function**. Now, let us focus in interpreting the `\(\beta_1\)` coefficient. #### Identity link For the identity link, the interpretation is straighforward. For one-unit increase in `\(x_1\)`, `\(\beta_1\)` dictates a constant difference in the outcome. `\(\Delta{y_i} = [\beta_0 + (\color{red}{x_{1i} + 1})\beta_1 + x_{2i}\beta_2] - (\beta_0 + x_{1i}\beta_1 + x_{2i}\beta_2)\)` `\(\Delta{y_i} = \beta_1\)` --- # Interpreting the `glm()` coefficients #### Logit link If a linear logistic model has been used with the two covariates `\(x_1\)` and `\(x_2\)`, we have the model: `$$log({\dfrac{p}{1-p}})=\beta_0 + x_{1i}\beta_1 + x_{2i}\beta_2$$` for a log odds of a positive response. We may also write that model as follows: `$$\dfrac{p}{1-p}=exp(\beta_0 + x_{1i}\beta_1 + x_{2i}\beta_2)$$` `$$Pr(yi) = \dfrac{exp(\beta_0 + x_{1i}\beta_1 + x_{2i}\beta_2)}{1 + exp{(\beta_0 + x_{1i}\beta_1 + x_{2i}\beta_2)}}$$` .center[Great! Do not give up!] --- # Interpreting the `glm()` coefficients #### Logit link Since the inverse link is nonlinear, the coefficient interpretation is difficult. Let us take a look what happens to the differences for a one-unit change to `\(x_1\)`: `$$\Delta{y_i} = \dfrac{\exp[\beta_0 + (\color{red}{x_{1i} + 1})\beta_1 + x_{2i}\beta_2]}{1 + \exp{[\beta_0 + (\color{red}{x_{1i} + 1})\beta_1 + x_{2i}\beta_2]}} - \dfrac{\exp[\beta_0 + x_{1i}\beta_1 + x_{2i}\beta_2]}{1 + \exp{[\beta_0 + x_{1i}\beta_1 + x_{2i}\beta_2]}}$$` $$\Delta{y_i} = exp(\beta_1) $$ As `\(x\)` increases by one unit, the odds increase by a factor of `\(\exp(\beta_1)\)`. --- # Interpreting the output ###### Getting back to our `logit.reg` model! .small[ ```r logit.reg # # Call: glm(formula = pa ~ WatrCont + Topo, family = binomial(link = "logit"), # data = mites) # # Coefficients: # (Intercept) WatrCont TopoHummock # 4.46440 -0.01581 2.09076 # # Degrees of Freedom: 69 Total (i.e. Null); 67 Residual # Null Deviance: 91.25 # Residual Deviance: 48.76 AIC: 54.76 ``` ] For a one-unit increase (or decrease) in water content, we can obtain the odds for the presence of mites. ```r exp(logit.reg$coefficient[2]) # WatrCont # 0.9843118 ``` --- # Interpreting the output .pull-left[ When odds = 1, the probability to observe the event `\(Y\)` is equal to the probability of *not* observing it (*i.e.* `\(p = 0.5\)`, so `\(0.5/(1-0.5) = 1\)`). When odds `\(< 1\)`, we have to take the inverse (_i.e._ 1 divided by the odds) to facilitate interpretation. The interpretation is then how **LESS** likely it is to observe the event of interest. For water content, the odds is `\(0.984\)`. The inverse is: `$$\dfrac{1}{0.984} = 1.0159$$` ] .pull-right[ This means that a one-unit increase in water content decreases the likelihood of observing _Galumna sp_. by `\(1.0159\)`. We can obtain a percentage change as below: `$$(1.0159 - 1) * 100 = 1.59%$$` There is a `\(1.59%\)` decrease in probability of observing _Galumna sp._ with a one-unit increase in water content. <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-52-1.png" width="288" style="display: block; margin: auto;" /> ] --- # Predictive power and goodness-of-fit Recall that the `\(R^2\)` value is a measure of how well the model explains the data. In simple linear models, `\(R^2\)` can be obtained as the square of the sample correlation equation coefficient ( `\(r\)` ) between the observed outcomes and the predictive values. -- In some of the generalized linear models, we take different approaches. We can obtain a `\(\text{pseudo-R}^2\)`, the analogue of the `\(R^2\)` for models fitted by maximum likelihood. We can calculate McFadden's (1973) `\(\text{pseudo-R}^2\)` in this way: `$$\text{pseudo-R}^2 = \frac{\text{null deviance - residual deviance}}{\text{null deviance}}$$` <br> ??? `\(\text{pseudo-R}^2\)` is the variance explained by the model. --- # Unit deviance, total deviance and null deviance The unit deviance is a measure of distance between `\(y\)` and `\(μ\)`. `$${\displaystyle d(y,y)=0}$$` `$${\displaystyle d(y,\mu )>0\quad \forall y\neq \mu }$$` ??? We will use the deviances to calculate the pseudo R2. -- The total deviance `\({\displaystyle D(\mathbf {y} ,{\hat {\boldsymbol {\mu }}})}\)` of a model with predictions `\({\hat {\boldsymbol {\mu }}}\)` of the observation `\(\mathbf {y}\)` is the sum of its unit deviances: `$${\displaystyle D(\mathbf {y} ,{\hat {\boldsymbol {\mu }}})=\sum _{i}d(y_{i},{\hat {\mu }}_{i})}$$` -- Now, the deviance of a model that has estimates `\({\hat {\mu }}=E[Y|{\hat {\theta }}_{0}]\)` can be defined by its **likelihood**: `$$D(y,{\hat {\mu }})=2{\Big (}\log {\big (}p(y\mid {\hat {\theta }}_{s}){\big )}-\log {\big (}p(y\mid {\hat {\theta }}_{0}){\big )}{\Big )}$$` where `\(\hat \theta_0\)` denotes the fitted values of the parameters in the reduced model, while `\(\hat \theta_s\)` denotes the fitted parameters for the saturated model. --- # Unit deviance, total deviance and null deviance The **residual deviance** is defined as 2 times the log-likelihood ratio of the full model compared to the reduced model. (The function below is exactly the same as above!) `$$D(y,{\hat {\mu }})=2{\Big (}\log {\big (}p(\text{saturated model}){\big )}-\log {\big (}p(\text{reduced model}){\big )}{\Big )}$$` -- And, the **null deviance** is defined 2 times the log-likelihood ratio of the full model compared to the null model (i.e. predictors are set to 1). `$$D(y,{\hat {\mu }})=2{\Big (}\log {\big (}p(\text{saturated model}){\big )}-\log {\big (}p(\text{null model}){\big )}{\Big )}$$` ??? Under some conditions, if the proposed model describes the data nearly as well as the saturated model, then asymptotically D ∼ χ 2 K−p, where K and p are the number of parameters in the saturated and proposed models, respectively. If the proposed model is poor, D will be larger than predicted by the χ2 K−p distribution. --- # Total deviance and null deviance In `R`, we can do this as follows: Let us compare the deviance of your model (residual deviance) to the deviance of a null model (null deviance). -- The **null deviance model** here is a model without explanatory variables. ```r null.model <- glm(response.variable ~ 1, family = binomial) ``` -- The **saturated (or full) deviance model** here is a model with all explanatory variables. ```r full.model <- glm(response.variable ~ ., family = binomial) ``` --- # Predictive power and goodness-of-fit In `R`, we can extract the residual and null deviances directly from the `glm object`: .small[ ```r objects(logit.reg) # [1] "aic" "boundary" "call" # [4] "coefficients" "contrasts" "control" # [7] "converged" "data" "deviance" # [10] "df.null" "df.residual" "effects" # [13] "family" "fitted.values" "formula" # [16] "iter" "linear.predictors" "method" # [19] "model" "null.deviance" "offset" # [22] "prior.weights" "qr" "R" # [25] "rank" "residuals" "terms" # [28] "weights" "xlevels" "y" ``` ] -- Using the deviance values available from the `logit.reg` object what is the pseudo `\(R^2\)` value? ??? Instruct participants to write some code based on the learning about deviance to manually calculate `\(R^2\)` and write their answer in the chat --- # Predictive power and goodness-of-fit In `R`, we can extract the residual and null deviances directly from the `glm object`: .small[ ```r objects(logit.reg) # [1] "aic" "boundary" "call" # [4] "coefficients" "contrasts" "control" # [7] "converged" "data" "deviance" # [10] "df.null" "df.residual" "effects" # [13] "family" "fitted.values" "formula" # [16] "iter" "linear.predictors" "method" # [19] "model" "null.deviance" "offset" # [22] "prior.weights" "qr" "R" # [25] "rank" "residuals" "terms" # [28] "weights" "xlevels" "y" ``` ```r pseudoR2 <- (logit.reg$null.deviance - logit.reg$deviance) / logit.reg$null.deviance pseudoR2 # [1] 0.4655937 ``` ] <br> .comment[Hence, the model explains 46.6% of the variability in the data.] --- # Predictive power and goodness-of-fit An adjusted McFadden's pseudo-R2, which penalizes for the number of predictors, can be calculated as below: .pull-left[ `\begin{equation} R^2_{adj} = 1 - \frac{logL(M) - K}{logL(M_{null})} \end{equation}` ] .pull-right[ where `\(K\)` corresponds to the additional number of predictors in relation to the null model. ] The goodness-of-fit of logistic regression models can be expressed by variants of pseudo-R2 statistics, such as Maddala (1983) or Cragg and Uhler (1970) measures. When talking about *logistic regressions*, low `\(R^2\)` values are often common. --- # Predictive power and goodness-of-fit The `DescTools::PseudoR2()` function computes several pseudo-R2. ```r logit.reg <- glm(pa ~ WatrCont + Topo, data = mites, family = binomial(link = "logit")) DescTools::PseudoR2(logit.reg, which = "all") # McFadden McFaddenAdj CoxSnell Nagelkerke AldrichNelson # 0.4655937 0.3998373 0.4549662 0.6245898 0.3776866 # VeallZimmermann Efron McKelveyZavoina Tjur AIC # 0.6674318 0.5024101 0.7064093 0.5114661 54.7623962 # BIC logLik logLik0 G2 # 61.5078819 -24.3811981 -45.6229593 42.4835224 ``` ??? One way to assess the goodness-of-fit of these models is to use the Hosmer-Lemeshow goodness-of-fit test which assess whether or not the observed event rates match expected event rates You can do this test with library(vcdExtra) HLtest(logit.reg) --- exclude: true ### Exercise 2: Performing a Hosmer-Lemeshow goodness-of-fit test One way to assess the goodness-of-fit of these models is to use the **Hosmer-Lemeshow goodness-of-fit test**. It roughly involves: 1. Dividing the data into groups of similar size; 2. Ordering on the predicted probability (or equivalently, the linear predictor); 3. Comparing the observed to expected number of positive responses in each group; 4. Performing a chi-squared test on the outcome. This test is assessing whether or not the observed event rates match expected event rates in subgroups of the model population. .pull-left[ ```r library(vcdExtra) HLtest(logit.reg) # Hosmer and Lemeshow Goodness-of-Fit Test # # Call: # glm(formula = pa ~ WatrCont + Topo, family = binomial(link = "logit"), # data = mites) # ChiSquare df P_value # 3.421693 8 0.9051814 ``` ] .pull-right[ ```r plot(HLtest(logit.reg)) ``` <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-58-1.png" width="432" style="display: block; margin: auto;" /> ] .comment[A non-significant value indicates an adequate fit!] --- # Challenge 2 ![:cube]() 1. Using the model created with `bacteria` dataset, assess goodness-of-fit and predictive power. 2. Think of how we could improve predictive power for this model. ??? Run this exercise as a breakout room activity with groups of 3-4 with a time limit of 5 minutes --- # Challenge 2 - Solution ![:cube]() 1\. Using the model created with `bacteria` dataset, assess goodness-of-fit and predictive power. ```r null.d <- model.bact2$null.deviance resid.d <- model.bact2$deviance bact.pseudoR2 <- (null.d - resid.d) / null.d bact.pseudoR2 # [1] 0.0624257 DescTools::PseudoR2(model.bact2, which = "all") # McFadden McFaddenAdj CoxSnell Nagelkerke AldrichNelson # 0.06242570 0.02562309 0.05981734 0.09529510 0.05809758 # VeallZimmermann Efron McKelveyZavoina Tjur AIC # 0.11689650 0.06151653 0.11098468 0.06275058 211.80606240 # BIC logLik logLik0 G2 # 225.38057258 -101.90303120 -108.68795256 13.56984272 ``` -- 2\. Think how predictive power could be improved for this model. *Adding informative explanatory variables could increase the explanatory power of the model.* *But, do not be afraid of non-significant results!* --- # Proportion data and GLM Sometimes, proportion data are more similar to logistic regression than you think... In discrete counts, we can, for instance, measure the number of presence of individuals in relation to the total number of populations sampled. We will thus obtain a proportional number of "success" in observing individuals by dividing the counts by the total counts. > In `glm()`, we have to provide *prior weights* if the response variable is the proportion of successes. --- # Proportion data and GLM Proportions can be modeled by providing both the number of "successes" and *prior weights* in the function ```r prop.reg <- glm(cbind(Galumna, totalabund - Galumna) ~ Topo + WatrCont, data = mites, family = binomial) ``` The weights can also be set explicitly in `glm()` ```r prop.reg2 <- glm(prop ~ Topo + WatrCont, data = mites, family = binomial, weights = totalabund) ``` --- exclude: true # Exercise 3 In `R`, we have to specify the number of times something happened and the number of times something did not happen: ```r prop.reg <- glm(cbind(Galumna, totalabund - Galumna) ~ Topo + WatrCont, data = mites, family = binomial) ``` ```r summary(prop.reg) ``` --- exclude: true # Exercise 3 .small[ ```r summary(prop.reg) # # Call: # glm(formula = cbind(Galumna, totalabund - Galumna) ~ Topo + WatrCont, # family = binomial, data = mites) # # Deviance Residuals: # Min 1Q Median 3Q Max # -1.4808 -0.9699 -0.6327 -0.1798 4.1688 # # Coefficients: # Estimate Std. Error z value Pr(>|z|) # (Intercept) -3.288925 0.422109 -7.792 6.61e-15 *** # TopoHummock 0.578332 0.274928 2.104 0.0354 * # WatrCont -0.005886 0.001086 -5.420 5.97e-08 *** # --- # Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 # # (Dispersion parameter for binomial family taken to be 1) # # Null deviance: 140.702 on 69 degrees of freedom # Residual deviance: 85.905 on 67 degrees of freedom # AIC: 158.66 # # Number of Fisher Scoring iterations: 5 ``` ] --- exclude: true # Exercise 3 We can also code the model directly with proportions: ```r prop.reg2 <- glm(prop ~ Topo + WatrCont, data = mites, family = binomial, weights = totalabund) ``` --- class: inverse, center, middle # GLM with count data --- # Modeling count data .large[What is count data?] First, import the `faramea.csv` into `R`. ```r faramea <- read.csv('data/faramea.csv', header = TRUE, stringsAsFactors = TRUE) ``` The number of trees of the species *Faramea occidentalis* was assessed in `\(43\)` quadrats in Barro Colorado Island (Panama). For each quadrat, environmental characteristics (such as, elevation and precipitation) were also recorded. Let us investigate the histogram of the frequency of *Faramea occidentalis*. --- # Modeling count data .large[What is count data?] Count data are often characterized by: - Positive values: you do not count **-7** individuals; - Integer values: you do not count **7.56** individuals; - Exhibits larger variance for larger values. <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-65-1.png" width="576" style="display: block; margin: auto;" /> ??? You can say that *on average*, you expect 7.56 individuals in a given situation. But you measure something *discrete*. --- # Modeling count data .large[How to model count data?] Does elevation influence the abundance of *F. occidentalis*? .pull-left[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-66-1.png" width="432" style="display: block; margin: auto;" /> ] .pull-right[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-67-1.png" width="432" style="display: block; margin: auto;" /> ] .small[What type of distribution best fits this data? 1. Bernoulli 2. Normal 3. Poisson 4. Binomial ] ??? This is a single choice poll question. The correct answer is 3. Poisson --- # Modeling count data .large[How to model count data?] Does elevation influence the abundance of *F. occidentalis*? .pull-left[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-68-1.png" width="432" style="display: block; margin: auto;" /> ] .pull-right[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-69-1.png" width="432" style="display: block; margin: auto;" /> ] .small[What type of distribution best fits this data? 1. Bernoulli 2. Normal 3. **Poisson** 4. Binomial ] ??? This is a single choice poll question. The correct answer is 3. Poisson --- # The Poisson distribution The Poisson distribution specifies the probability of a discrete random variable `\(Y\)` and is given by: `$$f(y, \,\mu)\, =\, Pr(Y = y)\, =\, \frac{\mu^y \times e^{-\mu}}{y!}$$` `$$E(Y)\, =\, Var(Y)\, =\, \mu$$` **Properties**: - `\(\mu\)` is the parameter of the Poisson distribution; - specifies the probably only for integer values; - probability for negative values is null ( `\(P(Y<0) = 0\)` ); - `\(Var(Y) = \mu\)` (allows for heterogeneity). --- # Poisson GLM behind the scenes A Poisson GLM will model the value of `\(\mu\)` as a function of different explanatory variables. <br> .center[**Three steps**] **Step 1.** We assume `\(Y_i\)` follows a Poisson distribution with mean and variance `\(\mu_i\)`. `$$Y_i ∼ Poisson(\mu_i)$$` `$$E(Y_i) = Var(Y_i) = \mu_i$$` `$$f(y_i, \, \mu_i) = \frac{\mu^{y_i}_i \times e^{-\mu_i}}{y!}$$` `\(\mu_i\)` corresponds to the expected number of individuals --- exclude:true # Poisson GLM behind the scenes **Step 2.** We specify the systematic part of the model just as in a linear model. `$$\underbrace{\alpha}_\text{Intercept} + \underbrace{\beta_1}_\text{slope of 'Elevation'} \times \text{Elevation}_i + \underbrace{\beta_2}_\text{slope of 'Precipitation'} \times \text{Precipitation}_i$$` -- exclude:true **Step 3.** The link between the mean of `\(Y_i\)` and the systematic part is a logarithmic function can be written as: `$$log(\mu_i) = \alpha + \beta_1 \times \text{Elevation}_i + \beta_2 \times \text{Precipitation}_i$$` .pull-left[ .center[or] `$$\mu_i = e^{ \alpha + \beta_1 \times \text{Elevation}_i + \beta_2 \times \text{Precipitation}_i}$$` .center[or] `$$\mu_i = e^{\alpha} \times e^{\beta_1^{\text{Elevation}_i}} \times e^{\beta_2^{\text{Precipitation}_i}}$$` ] .small[ This shows that the impact of each explanatory variable is multiplicative. Increasing Elevation by one increases μ by factor of exp( `\(\beta_\text{Elevation}\)` ). If `\(βj = 0\)` then `\(exp(βj) = 1\)` and `\(μ\)` is not related to `\(x_j\)`. If `\(βj > 0\)` then `\(μ\)` increases if `\(x_j\)` increases; if `\(βj < 0\)` then `\(μ\)` decreases if `\(x_j\)` increases. ] --- # Poisson GLM behind the scenes **Step 2.** We specify the systematic part (linear predictor) of the model just as in a linear model. `$$\underbrace{\alpha}_\text{Intercept} + \underbrace{\beta_1}_\text{slope of Elevation} \times \text{Elevation}_i$$` -- **Step 3.** The link between the mean of `\(Y_i\)` and the systematic part is a logarithmic function can be written as: .pull-left[ `$$log(\mu_i) = \alpha + \beta_1 \times \text{Elevation}_i$$` .center[or] `$$\mu_i = e^{ \alpha + \beta \times \text{Elevation}_i}$$` ] .small[ This shows that the impact of each explanatory variable is multiplicative. Increasing Elevation by one increases μ by factor of exp( `\(\beta_\text{Elevation}\)` ). If `\(βj = 0\)` then `\(exp(βj) = 1\)` and `\(μ\)` is not related to `\(x_j\)`. If `\(βj > 0\)` then `\(μ\)` increases if `\(x_j\)` increases; if `\(βj < 0\)` then `\(μ\)` decreases if `\(x_j\)` increases. ] --- # Fitting a Poisson GLM in `R` The function `glm()` allows you to specify a Poisson GLM. ```r glm.poisson = glm(Faramea.occidentalis ~ Elevation, data = faramea, family = poisson) ``` The `family` argument specifies the distribution and link function. <br> As with `lm()` you can access the outputs of the model using the function `summary()`: ```r summary(glm.poisson) ``` --- # Model summary .pull-left2[ .xsmall[ ```r summary(glm.poisson) # # Call: # glm(formula = Faramea.occidentalis ~ Elevation, family = poisson, # data = faramea) # # Deviance Residuals: # Min 1Q Median 3Q Max # -3.3319 -2.7509 -1.5451 0.1139 11.3995 # # Coefficients: # Estimate Std. Error z value Pr(>|z|) # (Intercept) 1.7687001 0.1099136 16.092 < 2e-16 *** # Elevation -0.0027375 0.0006436 -4.253 2.11e-05 *** # --- # Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 # # (Dispersion parameter for poisson family taken to be 1) # # Null deviance: 414.81 on 42 degrees of freedom # Residual deviance: 388.12 on 41 degrees of freedom # AIC: 462.01 # # Number of Fisher Scoring iterations: 10 ``` ] ] .pull-right2[ Estimates: Intercept = `\(\alpha\)` Elevation = `\(\beta\)` ] -- .pull-right2[ <br> What about the *null deviance* and the *residual deviance*? ] ??? Note: Point out that the residual deviance of 388.12 is much higher than residual degrees of freedom of 41 indicating overdispersion. --- # Parameter estimates In our model the unknown parameters are the intercept ( `\(\alpha\)` ) and the slope of elevation ( `\(\beta\)` ): `$$log(\mu_i) = 1.769 - 0.0027 \times \text{Elevation}_i$$` .center[or] `$$\mu_i = e^{1.769 - 0.0027 \times \text{Elevation}_i}$$` --- # The deviance Remember that to estimate the unknown parameter maximum likelihood estimation is used. The residual deviance is defined as twice the difference between the log-likelihood of a model that provides a perfect fit and the log-likelihood of our model. `$$\text{residual deviance} = 2 \, log(L(y;\,y)) - 2 \, log(L(y;\, \mu))$$` In a Poisson GLM, the residual deviance should be close to the residual degrees of freedom. .center[.alert[388.12 >> 41]] >Our residual deviance is much higher than the degrees of freedom of our model! --- # Overdispersion For a Poisson distribution, `\(var[y] = μ\)`. However, in practice the apparent variance of the data often exceeds `\(μ\)`, reflecting *overdispersion* in the model parameters. <br> Overdispersion arises because the mean `\(μ\)` innately varies, even when all the explanatory variables are fixed, or because the events that are being counted are positively correlated. <br> .center[*Are you able to think of a situation in biology that can cause overdispersion?*] <br> -- <br> **Why should we care about overdispersion?** Tests on the explanatory variables will generally appear to be more significant and confidence intervals for the parameters will be narrower than warranted by the data! --- # Overdispersion When the residual deviance is higher than the residual degrees of freedom we say that the model is **overdispersed**. We can calculate an overdispersion parameter ( `\(\phi\)` ): `$$\phi \, = \, \frac{\text{Residual deviance}}{\text{Residual degrees of freedom}}$$` Since the Poisson GLM assumes that `\(var[y] = μ\)`, we will need to find an alternative. .center[.large[**Solutions**]] .pull-left[ 1: Correct for overdispersion using a **quasi-Poisson** generalized linear model. ] .pull-right[ 2: Choose another distribution family, such as the **negative binomial**. ] --- # Quasi-Poisson GLM The variance of the distribution will account for **overdispersion**: `$$E(Y_i) = \mu_i$$` `$$Var(Y_I) = \phi \times \mu_i$$` where `\(\phi\)` (*phi*) is the dispersion parameter. The **systematic part** and the **link function** remain the same. Correcting for overdispersion will not affect parameter estimates, but will affect their **significance**. Further, the standard errors of the parameters are multiplied by `\(\sqrt{\phi}\)`. .alert[Some marginally significant p-values may no longer hold!] --- exclude: false # Fitting a quasi-Poisson GLM in `R` Create a new `glm.object` using the '`quasipoisson`' family or update the previous one: ```r glm.quasipoisson = glm(Faramea.occidentalis ~ Elevation, data = faramea, family=quasipoisson) glm.quasipoisson = update(glm.poisson, family = quasipoisson) ``` --- # Fitting a quasi-Poisson GLM in `R` .pull-left2[ .small[ ```r summary(glm.quasipoisson) # # Call: # glm(formula = Faramea.occidentalis ~ Elevation, family = quasipoisson, # data = faramea) # # Deviance Residuals: # Min 1Q Median 3Q Max # -3.3319 -2.7509 -1.5451 0.1139 11.3995 # # Coefficients: # Estimate Std. Error t value Pr(>|t|) # (Intercept) 1.768700 0.439233 4.027 0.000238 *** # Elevation -0.002738 0.002572 -1.064 0.293391 # --- # Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 # # (Dispersion parameter for quasipoisson family taken to be 15.96936) # # Null deviance: 414.81 on 42 degrees of freedom # Residual deviance: 388.12 on 41 degrees of freedom # AIC: NA # # Number of Fisher Scoring iterations: 10 ``` ]] .pull-right2[ **Same estimates but** .small[The standard errors of the parameters are multiplied by] `$$\sqrt{\phi} = 4$$` `0.0006436 * 4 = 0.00257` <- `\(\phi\)` <br> <- .small[AIC is not defined] ] --- # Fitting a quasi-Poisson GLM in R Try also deviance analysis to test for the effect of Elevation ```r null.model <- glm(Faramea.occidentalis ~ 1, data = faramea, family = quasipoisson) anova(null.model, glm.quasipoisson, test = "Chisq") # Analysis of Deviance Table # # Model 1: Faramea.occidentalis ~ 1 # Model 2: Faramea.occidentalis ~ Elevation # Resid. Df Resid. Dev Df Deviance Pr(>Chi) # 1 42 414.81 # 2 41 388.12 1 26.686 0.1961 ``` --- # Dispersion parameter .center[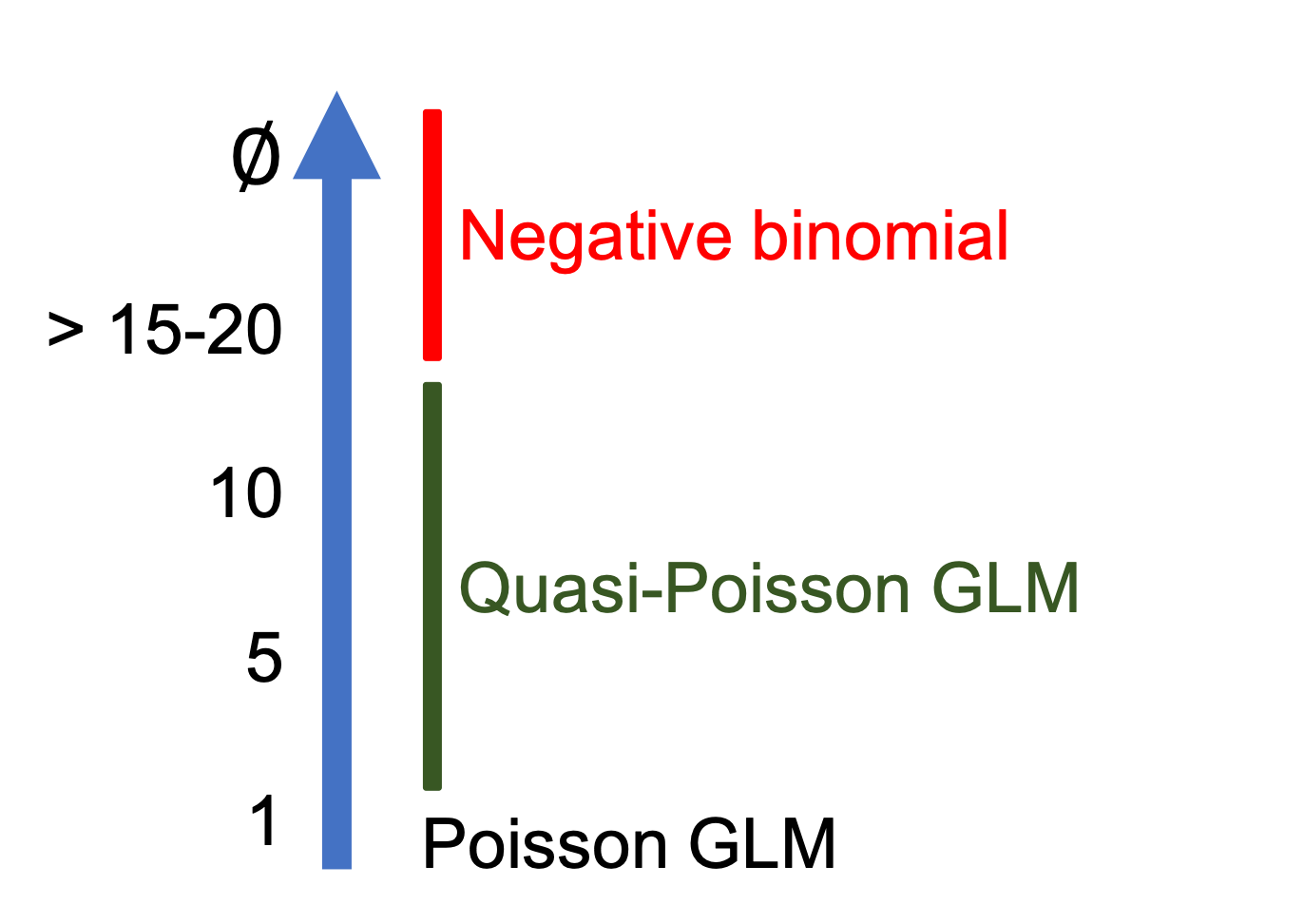] ??? A quasi-Poisson GLM can correct for some overdispersion but if too high a different distribution might be better --- # Negative binomial GLM Negative binomial GLMs are favoured when overdispersion is high: - It has **two parameters** `\(\mu\)` and `\(k\)`, where `\(k\)` controls for the dispersion parameter (smaller `\(k\)` indicates higher dispersion); - It corresponds to a combination of two distributions (**Poisson** and **Gamma**); - It assumes that the `\(Y_i\)` are Poisson distributed with the mean `\(\mu\)` assumed to follow a Gamma distribution! The predicted values follow as: `$$E(Y_i) = \mu_i$$` and the variance function: `$$Var(Y_i) = \mu_i + \frac{\mu^2_i}{k}$$` --- # Fitting a negative binomial in R NB is not in the `glm()` function so you need to install and charge the `MASS` package. ```r install.packages('MASS') ``` ```r glm.negbin <- glm.nb(Faramea.occidentalis ~ Elevation, data = faramea) ``` ```r summary(glm.negbin) ``` --- .pull-left2[ .small[ ``` # # Call: # glm.nb(formula = Faramea.occidentalis ~ Elevation, data = faramea, # init.theta = 0.2593107955, link = log) # # Deviance Residuals: # Min 1Q Median 3Q Max # -1.36748 -1.17564 -0.51338 -0.05226 2.25716 # # Coefficients: # Estimate Std. Error z value Pr(>|z|) # (Intercept) 2.369226 0.473841 5.00 5.73e-07 *** # Elevation -0.007038 0.002496 -2.82 0.00481 ** # --- # Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 # # (Dispersion parameter for Negative Binomial(0.2593) family taken to be 1) # # Null deviance: 41.974 on 42 degrees of freedom # Residual deviance: 36.343 on 41 degrees of freedom # AIC: 182.51 # # Number of Fisher Scoring iterations: 1 # # # Theta: 0.2593 # Std. Err.: 0.0755 # # 2 x log-likelihood: -176.5090 ``` ]] .pull-right2[ <br><br><br><br><br><br><br><br><br><br><br><br><br> `theta` `\(= k\)` ] --- # Plotting the final GLM model **Step 1** Plot the data and the use estimates of the parameters to draw model line `$$\mu_i = e^{2.369 - 0.007 \times Elevation_i}$$` Use `summary()` to get the parameters ```r summary(glm.negbin)$coefficients[1, 1] summary(glm.negbin)$coefficients[2, 1] ``` **Step 2** Use the standard errors to build the confidence interval ```r summary(glm.negbin)$coefficients[1, 2] summary(glm.negbin)$coefficients[2, 2] ``` `$$\text{Upper limit} = e^{[\alpha - 1.96 \times SE_{\alpha}] + [\beta - 1.96 \times SE_{\beta}] \times \text{Elevation}_i}$$` `$$\text{Upper limit} = e^{[\alpha + 1.96 \times SE_{\alpha}] + [\beta + 1.96 \times SE_{\beta}] \times \text{Elevation}_i}$$` --- .small[ ```r pp <- predict(glm.negbin, newdata = data.frame(Elevation = 1:800), se.fit = TRUE) linkinv <- family(glm.negbin)$linkinv ## inverse-link function pframe$pred0 <- pp$fit pframe$pred <- linkinv(pp$fit) sc <- abs(qnorm((1-0.95)/2)) ## Normal approx. to likelihood pframe <- transform(pframe, lwr = linkinv(pred0-sc*pp$se.fit), upr = linkinv(pred0+sc*pp$se.fit)) plot(faramea$Elevation, faramea$Faramea.occidentalis, ylab = 'Number of F. occidentalis', xlab = 'Elevation(m)') lines(pframe$pred, lwd = 2) lines(pframe$upr, col = 2, lty = 3, lwd = 2) lines(pframe$lwr, col = 2, lty = 3, lwd = 2) ``` ] <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-79-1.png" width="432" style="display: block; margin: auto;" /> --- # Challenge 3 ![:cube]() Use the `mites` dataset! Model the abundance of the species Galumna as a function of the substrate characteristics (water content `WatrCont` and density `SubsDens`) - Do you need to account for overdispersion? - Which covariates have a significant effect? - Select the best model! ```r mites <- read.csv("data/mites.csv", header = TRUE) ``` ??? This is a breakout activity with groups of 3-4 people and a time limit of 10 minutes. Don't forget to show tips on the next slide. --- # Challenge 3: Tips ![:cube]() Drop each term in turn and compare the full model with a nested model using the command: ```r drop1(MyGLM, test = "Chi") ``` Specify manually a nested model, call it for example MyGLM2, and use the command: ```r anova(MyGLM, MyGLM2, test = "Chi") ``` --- # Challenge 3: Solution ![:cube]() .small[ ```r # Poisson GLM glm.p = glm(Galumna~WatrCont+SubsDens, data=mites, family=poisson) # quasi-Poisson GLM glm.qp = update(glm.p,family=quasipoisson) # model selection drop1(glm.qp, test = "Chi") # Single term deletions # # Model: # Galumna ~ WatrCont + SubsDens # Df Deviance scaled dev. Pr(>Chi) # <none> 101.49 # WatrCont 1 168.10 31.711 1.789e-08 *** # SubsDens 1 108.05 3.125 0.07708 . # --- # Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ```r # or glm.qp2 = glm(Galumna~WatrCont, data=mites, family=quasipoisson) anova(glm.qp2, glm.qp, test="Chisq") ``` ] --- # Challenge 3: Solution ![:cube]() <br> .center[ <img src="workshop06-pres-en_files/figure-html/unnamed-chunk-83-1.png" width="432" style="display: block; margin: auto;" /> ] --- # Other distributions - **Logit transformation of the data** often used with `lm(m)` for percentages or proportions when the binomial distribution is not appropriate. When not selections from fixed quantities (e.g. percent cover, school grades, etc). - **Log-normal distribution in glm**, avoids having to log-transform the data. - **Gamma distribution**. Similar to log-normal, more versatile. - **Tweedie distribution**. Versatile family of distributions. Useful for data with a mix of zeros and positive values (not necessarily counts). - **Zero-inflated Poisson or zero-inflated negative binomial**. When the data comprise an excess number of zeros, that arise from a different process than the process that generates the counts. --- # Additional GLM resources **Books**: - B. Bolker (2009) Ecological Models and Data in R. Princeton University Press. - A. Zuur et al. (2009) Mixed Effects Models and Extensions in Ecology with R. Springer. **Articles** : - [Harrison et al. (2018), PeerJ, DOI 10.7717/peerj.4794 ](http://dx.doi.org/10.7717/peerj.4794) **Websites**: - GLMM for Ecologists (http://glmm.wikidot.com) .small[.comment[A great website on GLMM with a Q&A section!]] --- class: inverse, center, bottom # Thank you for attending this workshop!