Workshop 1: Introduction to R
QCBS R Workshop Series
Québec Centre for Biodiversity Science
Learning Objectives
1. Recognize and use R and RStudio;
2. Use R as a calculator;
3. Manipulate objects in R;
4. Install and use R packages and functions;
5. Get help.
1. Recognizing and using R and RStudio
Introduction
What is R?
Ris a free an open-source programming language and environment.
- It is designed for data analysis, graphical display and data simulations.
- It is one of the world's leading statistical programming environments.

Why should I become an useR?
Ris free, open source: built for you and for everyone;Ris popular: a large enganged user-base fosters the continued development and the maintainance of statistical tools;Ris powerful- You can program complex simulations
- Use it on high performance clusters
Rsupports extensionsRruns on most operating systemsRconnects with other languages:C++,Java,Python,Julia,Stanand more!

Why should I become an useR?
An example of a workflow to analyze data without R.
Why use R?
An example of a workflow to analyze data without R.
R allows you to do a lot without needing to use other programs.
Why use R?
- More and more scientists use it every year!
- As of October 2020, there are more than 16000 packages registered within the Comprehensive R Archive Network (CRAN) (and thousands more within Github repositories)!
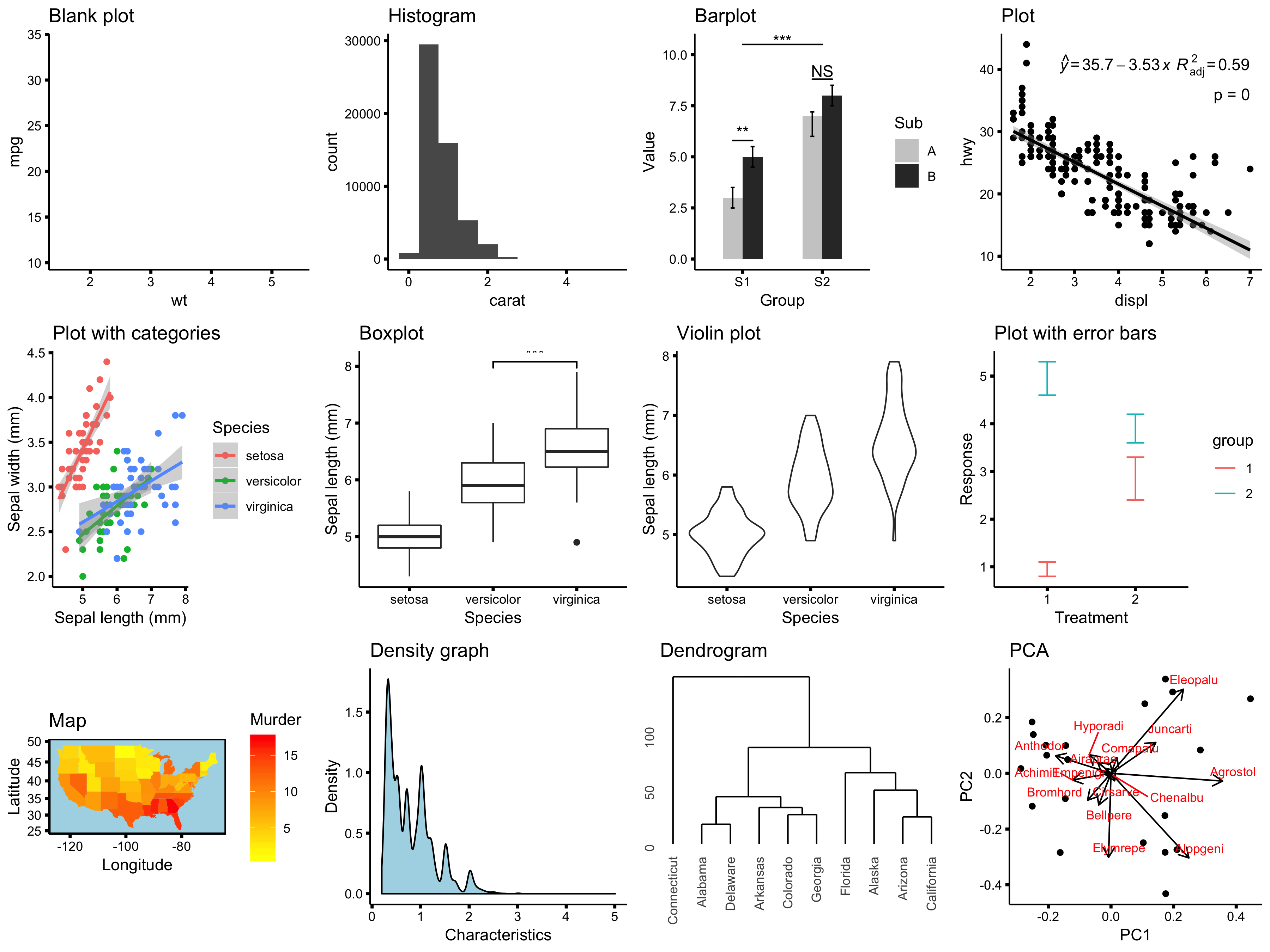
A lot of features: customizable graphs
All of these graphs were made in R!
And what about RStudio?
RStudio is the most used Integrated Development Environment (IDE) for R.
It includes a console, a syntax-highlighting editor that supports direct code execution with tools for plotting, history, debugging and workspace management.
It integrates with R (and other programming languages) to provide a lot of useful features:
RStudio supports authoring HTML, PDF, Word and presentation documents
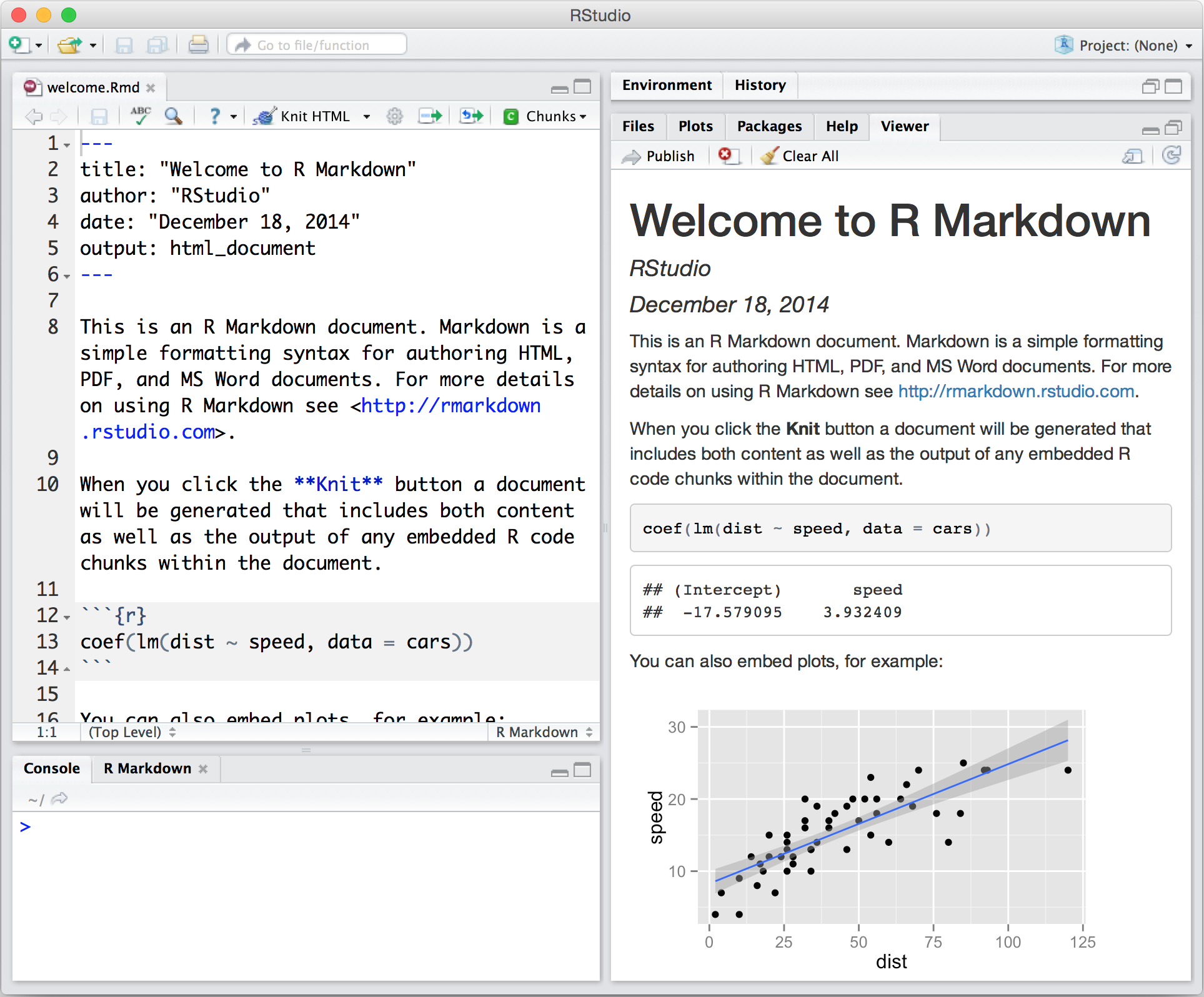
RStudio supports version control with Git (directly to Github) and Subversion
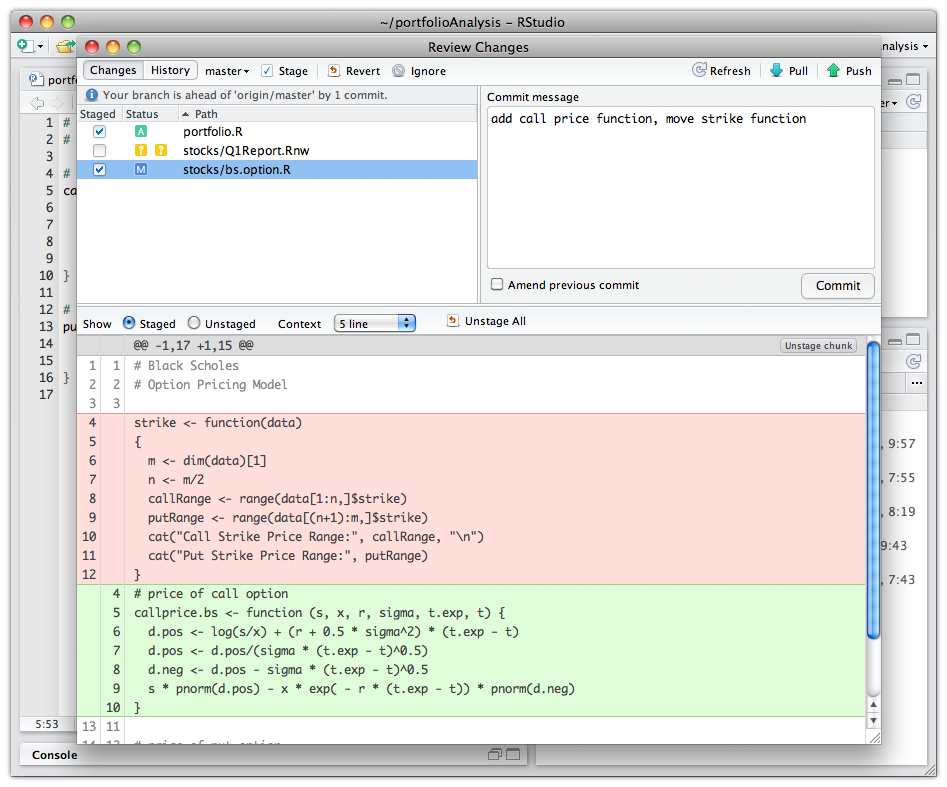
And what about RStudio?
RStudio is the most used Integrated Development Environment (IDE) for R.
It includes a console, a syntax-highlighting editor that supports direct code execution with tools for plotting, history, debugging and workspace management.
It integrates with R (and other programming languages) to provide a lot of useful features:
RStudio make it easy to start new or find existing projects
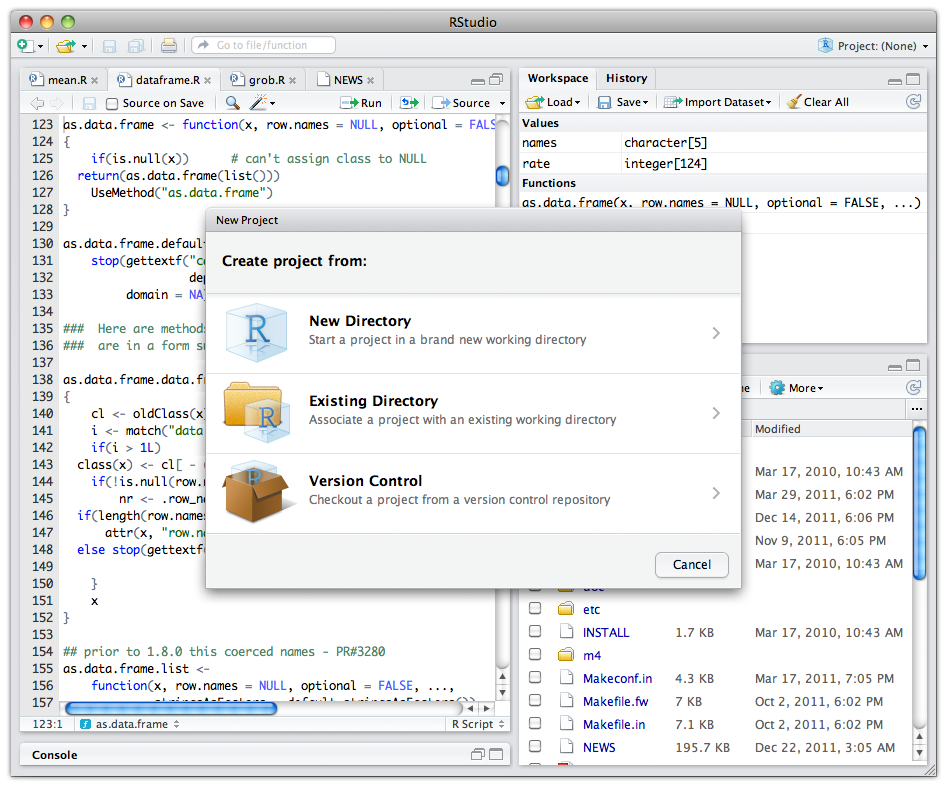
RStudio supports interactive graphics with Shiny and ggvis
And what about RStudio?
RStudio is the most used Integrated Development Environment (IDE) for R.
It includes a console, a syntax-highlighting editor that supports direct code execution with tools for plotting, history, debugging and workspace management.
It integrates with R (and other programming languages) to provide a lot of useful features:
RStudio make it easy to start new or find existing projects

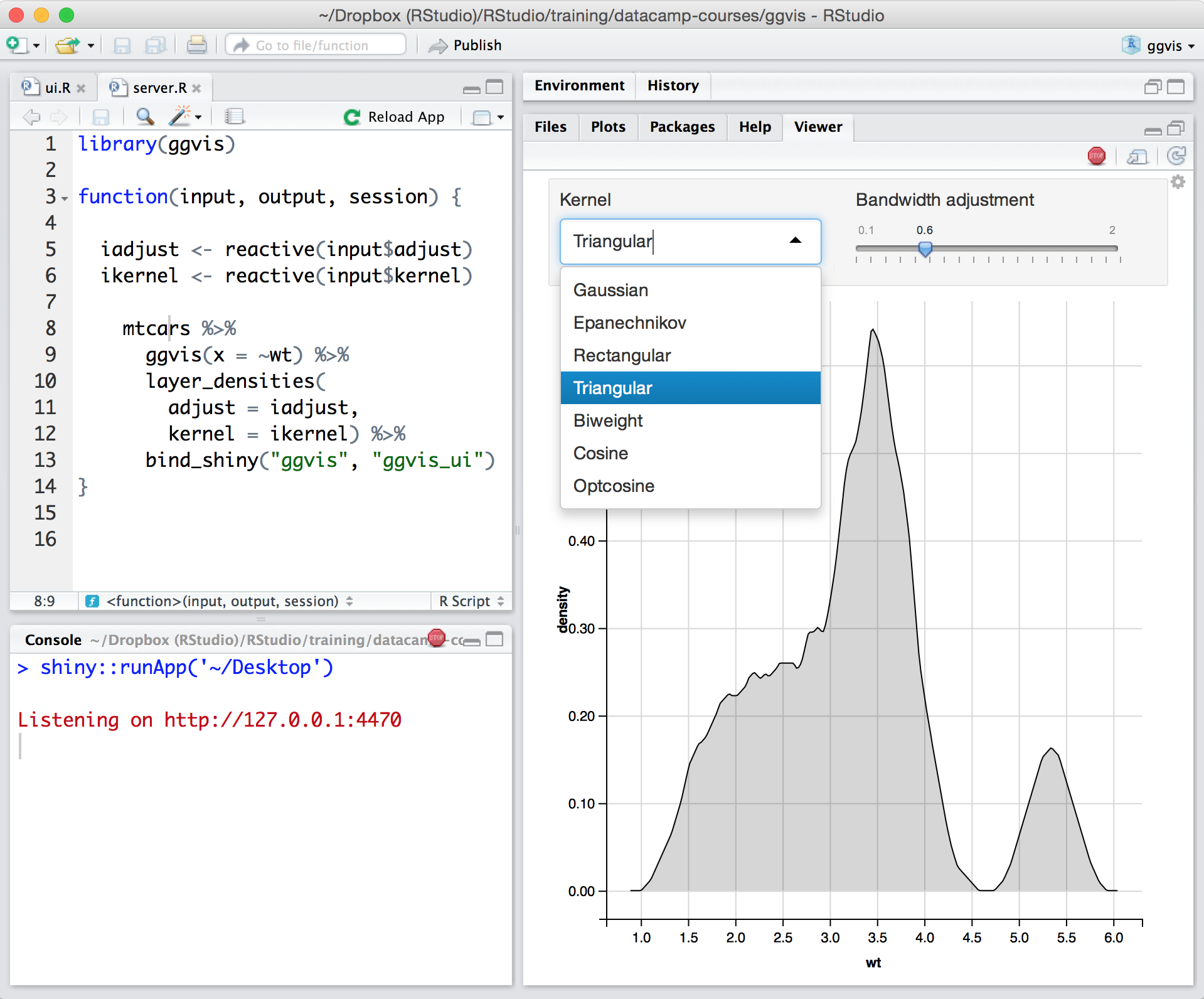
RStudio supports interactive graphics with Shiny and ggvis

There are other IDE for R: Atom, Visual Studio, Jupyter notebook and Jupyter lab!
Challenge
- Throughout these workshops, challenges will be indicated by Rubik cubes.
- Sometimes, you will be expected to collaborate with other participants!
- Do not hesitate to ask questions!

First Challenge
Open RStudio.
Let us do this one together!

Here, the workshop instructor may minimize the presentation and show his RStudio window and point out all View panes being displayed, briefly explaining each one of them.
Be sure to adjust the font size and scale of the window according to the presentation being given (remote presentations allow for smaller font size).
Note for Windows users
If the restriction unable to write on disk appears when you attempt to open RStudio or to install a package.
Do not worry!
We have the solution!
- Close the application.
- Right-click on your RStudio icon and click on "Execute as Administrator". This will provide file and directory writing rights to RStudio.
The RStudio Interface
When you open RStudio for the first time, the screen will be divided across three main Pane groups:
- Console, Terminal, Job group;
- Environment, History, Connections group;
- Files, Plot, Packages, Help, Viewer panes; and,
- Script pane group.
Once you Open a Script or Create a New Script (File > New File > R Script or Ctrl/Cmd + Shit + N), the fourth panel will appear!

Most of the action happens here!
The RStudio Console
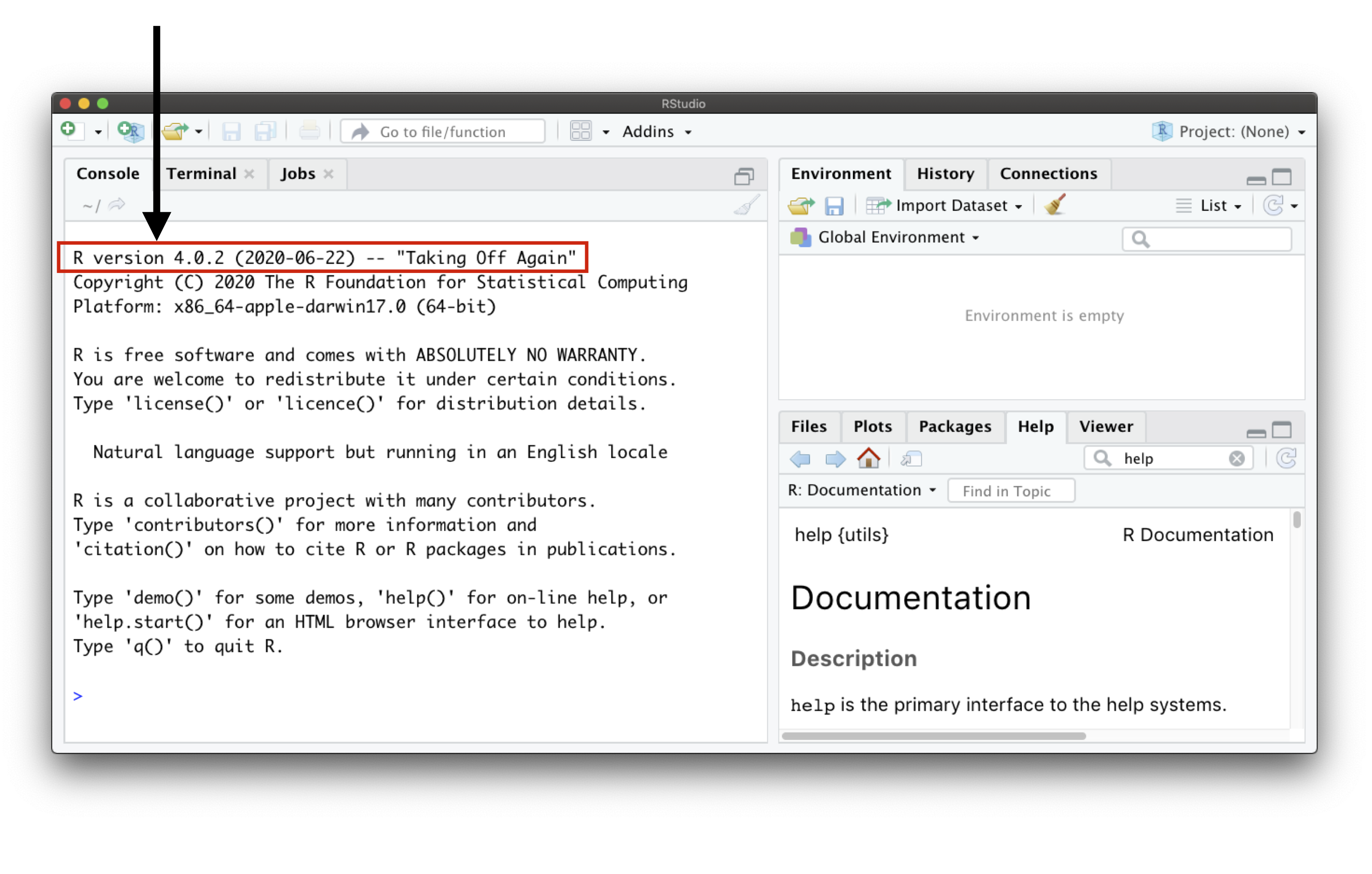
Usually, the first text you see within the Console pane is the
Rversion RStudio is using.The Console is the place where
Ris waiting for you to tell it what to do, and is where it will communicate with you, showing the outcome of your command.Whenever
Ris ready to accept commands, it will show a>prompt.
Reading the Console
Text in the console typically looks like this:
output# [1] "This is the output"
Remember that one must write the command in front of the > prompt and then press "Return" it to run.
What does the square brackets [ ] within the output mean?
Reading the Console
Text in the console typically looks like this:
output# [1] "This is the output"
Remember that one must write the command in front of the > prompt and then press "Return" it to run.
What does the square brackets [ ] within the output mean?
The numbers within the brackets help you to locate the position of elements within the output.
seq(1, 100, by = 2)# [1] 1 3 5 7 9 11 13 15 17 19 21 23 25 27 29 31 33 35 37 39 41 43 45 47 49# [26] 51 53 55 57 59 61 63 65 67 69 71 73 75 77 79 81 83 85 87 89 91 93 95 97 99Show the participants how the number between the square brackets indicates position of the elements.
Error and Warning
Often, the Console will output Errors and Warning messages.
Warning message
x <- c("2", -3, "end", 0, 4, 0.2)as.numeric(x)# Warning: NAs introduced by coercion# [1] 2.0 -3.0 NA 0.0 4.0 0.2- Cautions users about an action, but still executes the function.
- There might be an issue with the input and/or the output.
Error message
x*10# Error in x * 10: non-numeric argument to binary operator- Informs the user that there is a problem that prevents the command from running.
- One needs to solve the issue in order to carry on.
Google is your best friend in solving Errors or Warnings!
2. Using R as a calculator
Basic operations
Arithmetic Operators
- Additions and Subtractions
1 + 1# [1] 210 - 1# [1] 9Arithmetic Operators
- Additions and Subtractions
1 + 1# [1] 210 - 1# [1] 9- Multiplications and Divisions
2 * 2# [1] 48 / 2# [1] 4Arithmetic Operators
- Additions and Subtractions
1 + 1# [1] 210 - 1# [1] 9- Multiplications and Divisions
2 * 2# [1] 48 / 2# [1] 4- Exponents
2^3# [1] 8Challenge 
Use R to calculate the following equation:
2+16∗24−56
Hint: The * symbol is used to multiply.
Challenge: Solution 
Use R to calculate the following equation:
2+16∗24−56
It would look like this in R:
2 + 16 * 24 - 56# [1] 330Challenge 
Use R to calculate the following equation:
2+16∗24−56/(2+1)−457
Hint: Think about the order of the operation.
Challenge: Solution 
Use R to calculate the following equation:
2+16∗24−56/(2+1)−457
It would look like this in R:
2 + 16 * 24 - 56 / (2 + 1) - 457# [1] -89.66667Note that R respects the order of the operations
Still using R for arithmetic operations
What is the area of a circle with a radius of 5 cm?
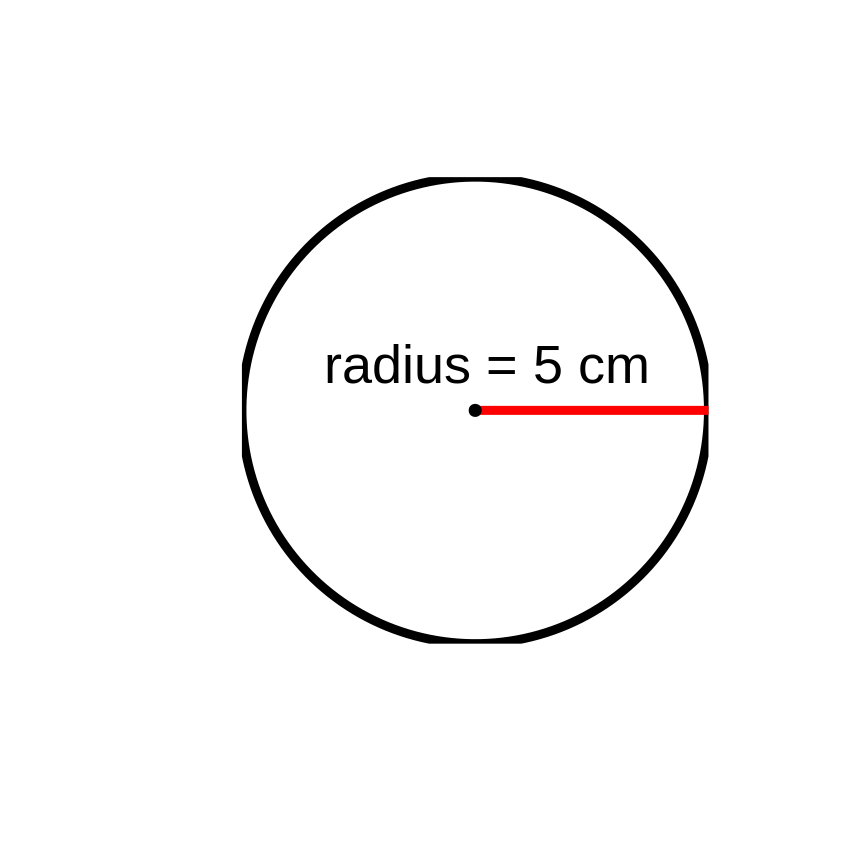
Areacircle=π×r2
Still using R for arithmetic operations
What is the area of a circle with a radius of 5 cm?
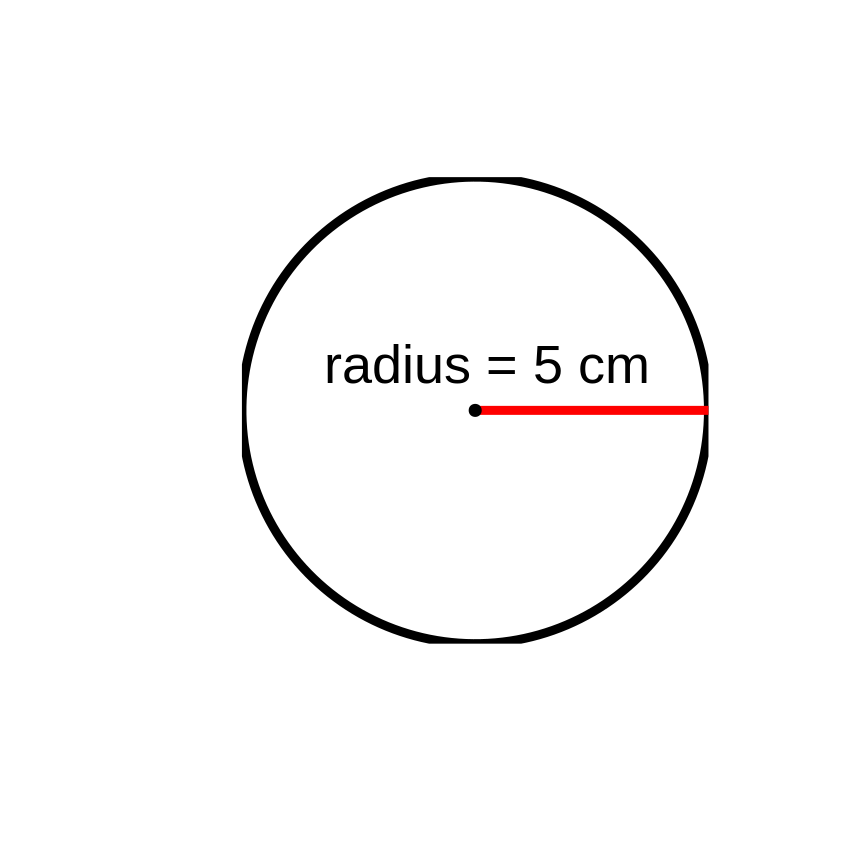
Areacircle=π×r2
3.1416 * 5^2# [1] 78.54Still using R for arithmetic operations
What is the area of a circle with a radius of 5 cm?

Areacircle=π×r2
3.1416 * 5^2# [1] 78.54But... R has built-in constants!
You can find them by typing ? and Constants (as in ?Constants) and executing it! What is the one for π?
We can then write and execute this:
pi * 5^2# [1] 78.53982You have just ran a command preceeded by ?. What happened?
A tip
We can use the ↑ and ↓ arrow keys to retrieve commands previously run.
Make sure your cursor is blinking in front of the > prompt and give it a try!

3. Manipulating objects in R
R: an object-oriented environment
You can assign information to named objects using the assignment operator <-.
The information is assigned to the name that is pointed by the assignment operator <-.
See the examples below:
money_talks <- "ACDC"money_talks# [1] "ACDC"9 -> my_birthday_monthmy_birthday_month# [1] 9Careful! There is no space between the less than (<) and minus (-) signs.
R: an object-oriented environment
You can assign information to named objects using the assignment operator <-.
The information is assigned to the name that is pointed by the assignment operator <-.
See the examples below:
money_talks <- "ACDC"money_talks# [1] "ACDC"9 -> my_birthday_monthmy_birthday_month# [1] 9Careful! There is no space between the less than (<) and minus (-) signs.
One can assign values using = instead of the <- operator. We caution against using = to assign values to objects because the = is allowed at the top level only and also determines subexpressions.
Naming objects: a few rules
Objects names can only include:
| Type | Symbol |
|---|---|
| Letters | a-z A-Z |
| Numbers | 0-9 |
| Period | . |
| Underscore | _ |
- Objects names must always begin with a letter.
Ris case-sensitive:Data_1is different thandata_1.- You cannot use special characters (
@,/,#, etc.). - Object names must be unique:
data_1 <- 1will overwrite any previously objects nameddata_1
Good practice when naming objects and writing code
Short and explicit names are preferred.
- Naming a variable
varis not very informative.
You can separate words within a name using underscores ( _ ) or dots ( . ).
avg_richnessoravg.richnessare easier to read thanavgrichness.
Avoid using names of existing functions or constants (e.g., c, table, T, matrix)
Good practice when naming objects and writing code
Short and explicit names are preferred.
- Naming a variable
varis not very informative.
You can separate words within a name using underscores ( _ ) or dots ( . ).
avg_richnessoravg.richnessare easier to read thanavgrichness.
Avoid using names of existing functions or constants (e.g., c, table, T, matrix)
Add spaces around operators (=, +, -, <-, etc.) to make the code more readable.
Always put a space after a comma, and never before (like in regular English).
Preferred
mean_x <- (2 + 6) / 2mean_x# [1] 4Not preferred
meanx<-(2+6)/2meanx# [1] 4Challenge 
Create an object with a name (of your choice) that starts with a number. What happens?
Challenge: Solution 
Create an object with a name (of your choice) that starts with a number. What happens?
Creating an object name that starts with a number returns the following error:
Error: unexpected symbol in "your object name"Challenge 
Create an object with a value of 1 + 1.718282 (e or Euler's number) and name it euler_value.
Challenge: Solution 
Create an object with a value of 1 + 1.718282 (e or Euler's number) and name it euler_value.
euler_value <- 1 + 1.718282euler_value# [1] 2.718282Challenge: Solution 
Create an object with a value of 1 + 1.718282 (e or Euler's number) and name it euler_value.
euler_value <- 1 + 1.718282euler_value# [1] 2.718282What has happened in your RStudio window when you created this object?
The presenter should not here that the object will appear in the RStudio Environment.
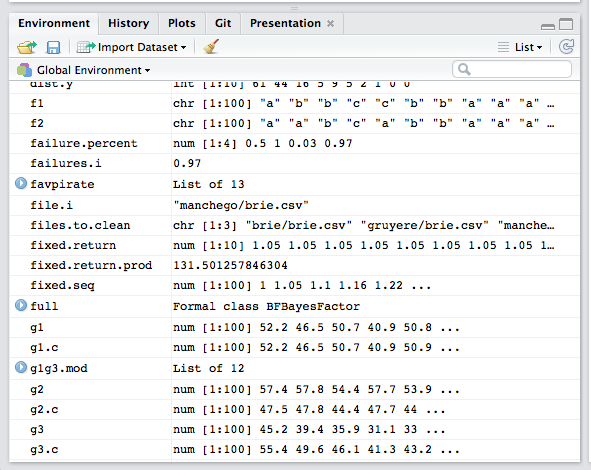
The RStudio Environment
The Environment panel shows you all the objects you have defined in your current workspace.
Tip
You can use the Tab key to auto-complete commands.
This helps preventing spelling errors
Let us try it!
Try writing eul or abb in front of the > prompt and press Tab.
If more than one element appears, you can use the arrow keys ( ↑ ↓ ) and press "Return" or use your mouse to select the correct one.
3. Manipulating objects in R
Data types and structure
Core data types in R
Data types define how the values are stored in R.
We can obtain the type and mode of an object using the functions typeof(). The core data types are:
Numeric-type with integer and double values
(x <- 1.1)# [1] 1.1typeof(x)# [1] "double"(y <- 2L)# [1] 2typeof(y)# [1] "integer"Core data types in R
Data types define how the values are stored in R.
We can obtain the type and mode of an object using the functions typeof(). The core data types are:
Numeric-type with integer and double values
(x <- 1.1)# [1] 1.1typeof(x)# [1] "double"(y <- 2L)# [1] 2typeof(y)# [1] "integer"Character-type (always between " ")
z <- "You are becoming very good in this!"typeof(z)# [1] "character"Core data types in R
Data types define how the values are stored in R.
We can obtain the type and mode of an object using the functions typeof(). The core data types are:
Numeric-type with integer and double values
(x <- 1.1)# [1] 1.1typeof(x)# [1] "double"(y <- 2L)# [1] 2typeof(y)# [1] "integer"Character-type (always between " ")
z <- "You are becoming very good in this!"typeof(z)# [1] "character"Logical-type
t <- TRUEtypeof(t)# [1] "logical"f <- FALSEtypeof(f)# [1] "logical"The presenter might be asked about running the entire line within (). This is the same as x <-2; x. We have done that so it fits.
Data structure in R: scalars
Until this moment, we have create objects that had just one element inside them:
x <- 1.1x# [1] 1.1euler_value <- 1 + 1.718282euler_value# [1] 2.718282An object that has just a single value or unit like a number or a text string is called a scalar.
a <- 100b <- 3 / 100c <- (a + b) / bd <- "species"e <- "genus"f <- "When is the next pause again?"Data structure in R: scalars
Until this moment, we have create objects that had just one element inside them:
x <- 1.1x# [1] 1.1euler_value <- 1 + 1.718282euler_value# [1] 2.718282An object that has just a single value or unit like a number or a text string is called a scalar.
a <- 100b <- 3 / 100c <- (a + b) / bd <- "species"e <- "genus"f <- "When is the next pause again?"
By creating combinations of scalars, we can create data with different structures in R. We are getting there!
Data structure in R: vectors
A vector object is just a combination of several scalars stored as a single object.
Like scalars, vectors can be of numeric-, logical-, character-types, but never a mix of them!
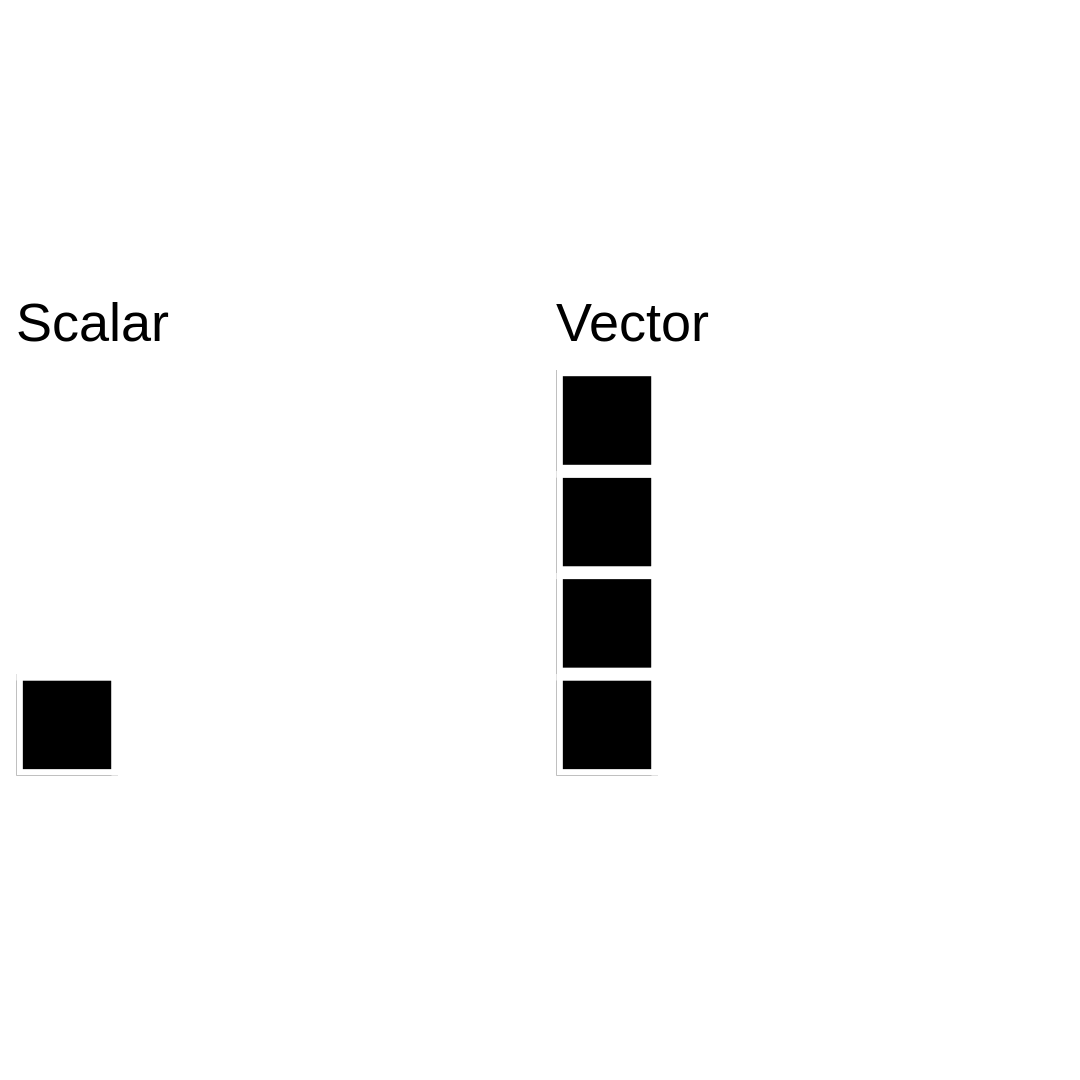
Data structure in R: vectors
A vector object is just a combination of several scalars stored as a single object.
Like scalars, vectors can be of numeric-, logical-, character-types, but never a mix of them!
There are many ways to create vectors in R. Here are some we are going to see:
| Function | Example | Result |
|---|---|---|
c(a, b, ...) |
c(1, 3, 5, 7, 9) |
1, 3, 5, 7, 9 |
a:b |
1:5 |
1, 2, 3, 4, 5 |
seq(from, to, by, length.out) |
seq(from = 0, to = 6, by = 2) |
0, 2, 4, 6 |
rep(x, times, each, length.out) |
rep(c(7, 8), times = 2, each = 2) |
7, 7, 8, 8, 7, 7, 8, 8 |
Creating vectors with c()
The c() function (c stands for concatenate, meaning bring them together) combines several scalars as arguments, which are separated by commas, and returns a vector containing them:
vector <- c(value1, value2, ...)Let us use the c() function to create vectors of different types:
Numeric vector
num_vector <- c(1, 4, 32, -76, -4)num_vector# [1] 1 4 32 -76 -4Character vector
char_vector <- c("blue", "red", "green")char_vector# [1] "blue" "red" "green"Logical vector
bool_vector <- c(TRUE, TRUE, FALSE) # or c(T, T, F)bool_vector# [1] TRUE TRUE FALSECreating vectors of sequential values: a:b, seq(), rep()
The a:b takes two numeric scalars a and b as arguments, and returns a vector of numbers from the starting point a to the ending point b, in steps of 1 unit:
1:8# [1] 1 2 3 4 5 6 7 87.5:1.5# [1] 7.5 6.5 5.5 4.5 3.5 2.5 1.5Creating vectors of sequential values: a:b, seq(), rep()
The a:b takes two numeric scalars a and b as arguments, and returns a vector of numbers from the starting point a to the ending point b, in steps of 1 unit:
1:8# [1] 1 2 3 4 5 6 7 87.5:1.5# [1] 7.5 6.5 5.5 4.5 3.5 2.5 1.5seq() allows us to create a sequence, like a:b, but also allows us to specify either the size of the steps (the by argument), or the total length of the sequence (the length.out argument):
seq(from = 1, to = 10, by = 2)# [1] 1 3 5 7 9seq(from = 20, to = 2, by = -2)# [1] 20 18 16 14 12 10 8 6 4 2Creating vectors of sequential values: a:b, seq(), rep()
The a:b takes two numeric scalars a and b as arguments, and returns a vector of numbers from the starting point a to the ending point b, in steps of 1 unit:
1:8# [1] 1 2 3 4 5 6 7 87.5:1.5# [1] 7.5 6.5 5.5 4.5 3.5 2.5 1.5seq() allows us to create a sequence, like a:b, but also allows us to specify either the size of the steps (the by argument), or the total length of the sequence (the length.out argument):
seq(from = 1, to = 10, by = 2)# [1] 1 3 5 7 9seq(from = 20, to = 2, by = -2)# [1] 20 18 16 14 12 10 8 6 4 2rep() allows you to repeat a scalar (or vector) a specified number of times, or to a desired length:
rep(x = 1:3, each = 2, times = 2)# [1] 1 1 2 2 3 3 1 1 2 2 3 3rep(x = c(1, 2), each = 3)# [1] 1 1 1 2 2 2The presenter should point out the differences between the rep example 1 and example 2, where a vector can be used inside the argument.
Challenge 
Let's practice:
- Create a vector containing the first 5 odd numbers, starting from 1
- Name it
odd_n - You can use any of the previous functions we have previously learned!
Challenge: Solution 
Let's practice:
- Create a vector containing the first 5 odd numbers, starting from 1
- Name it
odd_n - You can use any of the previous functions we have previously learned!
Solution:
odd_n <- c(1, 3, 5, 7, 9)or
odd_n <- seq(from = 1, to = 9, by = 2)odd_n# [1] 1 3 5 7 9Operations using vectors
Let us begin with the following objects:
x <- c(1:5)y <- 6
Remember that the colon symbol : combines all values between the first and the second number in steps of 1. c(1:5) or 1:5 is equivalent to c(1, 2, 3, 4, 5)
Operations using vectors
Let us begin with the following objects:
x <- c(1:5)y <- 6
Remember that the colon symbol : combines all values between the first and the second number in steps of 1. c(1:5) or 1:5 is equivalent to c(1, 2, 3, 4, 5)
What happens when we add and multiply the two objects together?
x + y# [1] 7 8 9 10 11x * y# [1] 6 12 18 24 30Operations using vectors
Let us begin with the following objects:
x <- c(1:5)y <- 6
Remember that the colon symbol : combines all values between the first and the second number in steps of 1. c(1:5) or 1:5 is equivalent to c(1, 2, 3, 4, 5)
What happens when we add and multiply the two objects together?
x + y# [1] 7 8 9 10 11x * y# [1] 6 12 18 24 30
Excellent! We have learned a lot! How about a short break?
Can you guess what our next topic is?
Data structure in R: matrices
We have learned that scalars contain one element, and that vectors contain more than one scalar of the same type!
Matrices are nothing but a bunch of vectors stacked together!
While vectors have one dimension, matrices have two dimensions, determined by rows and columns.
Finally, like vectors and scalars matrices can contain only one type of data: numeric, character, or logical.

Creating matrices using matrix(), cbind(), rbind()
There are many ways to create your own matrix. Let us start with a simple one:
matrix(data = 1:10, nrow = 5, ncol = 2)# [,1] [,2]# [1,] 1 6# [2,] 2 7# [3,] 3 8# [4,] 4 9# [5,] 5 10matrix(data = 1:10, nrow = 2, ncol = 5)# [,1] [,2] [,3] [,4] [,5]# [1,] 1 3 5 7 9# [2,] 2 4 6 8 10Creating matrices using matrix(), cbind(), rbind()
There are many ways to create your own matrix. Let us start with a simple one:
matrix(data = 1:10, nrow = 5, ncol = 2)# [,1] [,2]# [1,] 1 6# [2,] 2 7# [3,] 3 8# [4,] 4 9# [5,] 5 10matrix(data = 1:10, nrow = 2, ncol = 5)# [,1] [,2] [,3] [,4] [,5]# [1,] 1 3 5 7 9# [2,] 2 4 6 8 10We can also combine multiple vectors using cbind() and rbind():
nickname <- c("kat", "gab", "lo")animal <- c("dog", "mouse", "cat")rbind(nickname, animal)# [,1] [,2] [,3] # nickname "kat" "gab" "lo" # animal "dog" "mouse" "cat"cbind(nickname, animal)# nickname animal # [1,] "kat" "dog" # [2,] "gab" "mouse"# [3,] "lo" "cat"Operations with matrices
Similarly as in the case of vectors, operations with matrices work just fine:
(mat_1 <- matrix(data = 1:9, nrow = 3, ncol = 3))# [,1] [,2] [,3]# [1,] 1 4 7# [2,] 2 5 8# [3,] 3 6 9(mat_2 <- matrix(data = 9:1, nrow = 3, ncol = 3))# [,1] [,2] [,3]# [1,] 9 6 3# [2,] 8 5 2# [3,] 7 4 1The product of the matrices is:
mat_1 * mat_2# [,1] [,2] [,3]# [1,] 9 24 21# [2,] 16 25 16# [3,] 21 24 9Challenge 
It is your time to get your hands dirty!
- Create an object containing a matrix with 2 rows and 3 columns, with values from 1 to 6, sorted per column.
- Create another object with a matrix with 2 rows and 3 columns, with the names of six animals you like.
- Create a third object with 4 rows and 2 columns:
- in the first column, include the numbers from 2 to 5; and,
- in the second column, include the first name of participants in this workshop.
- Compare them and tell us what differences have you detected (despite their values).
Remember that text strings must always be surrounded by quote marks (" ").
Remember that values or arguments must be separated by commas if they are inside a function, e.g. c("one", "two", "three").
Challenge 
It is your time to get your hands dirty!
- Create an object containing a matrix with 2 rows and 3 columns, with values from 1 to 6, sorted per column.
- Create another object with a matrix with 2 rows and 3 columns, with the names of six animals you like.
(step_1 <- matrix(data = 1:6, nrow = 2, ncol = 3))# [,1] [,2] [,3]# [1,] 1 3 5# [2,] 2 4 6(step_2 <- matrix( data = c("cheetah", "tiger", "ladybug", "deer", "monkey", "crocodile"), nrow = 2, ncol = 3))# [,1] [,2] [,3] # [1,] "cheetah" "ladybug" "monkey" # [2,] "tiger" "deer" "crocodile"Challenge 
It is your time to get your hands dirty!
- Create a third object with 4 rows and 2 columns:
- in the first column, include the numbers from 2 to 5; and,
- in the second column, include the first name of participants in this workshop.
- Compare them and tell us what differences have you detected (despite their values).
step_1# [,1] [,2] [,3]# [1,] 1 3 5# [2,] 2 4 6step_2# [,1] [,2] [,3] # [1,] "cheetah" "ladybug" "monkey" # [2,] "tiger" "deer" "crocodile"step_3 <- cbind(c(2:5), c("linley", "jessica", "joe", "emma"))step_3# [,1] [,2] # [1,] "2" "linley" # [2,] "3" "jessica"# [3,] "4" "joe" # [4,] "5" "emma"The presenter here should mention that matrices being formed by vectors or scalars with multiple data types become characters. This will be a window to introduce data frames to the participants.
Data structure in R: data frames
Differently than a matrix, a data frame can contain numeric, character, and logical columns (or vectors).
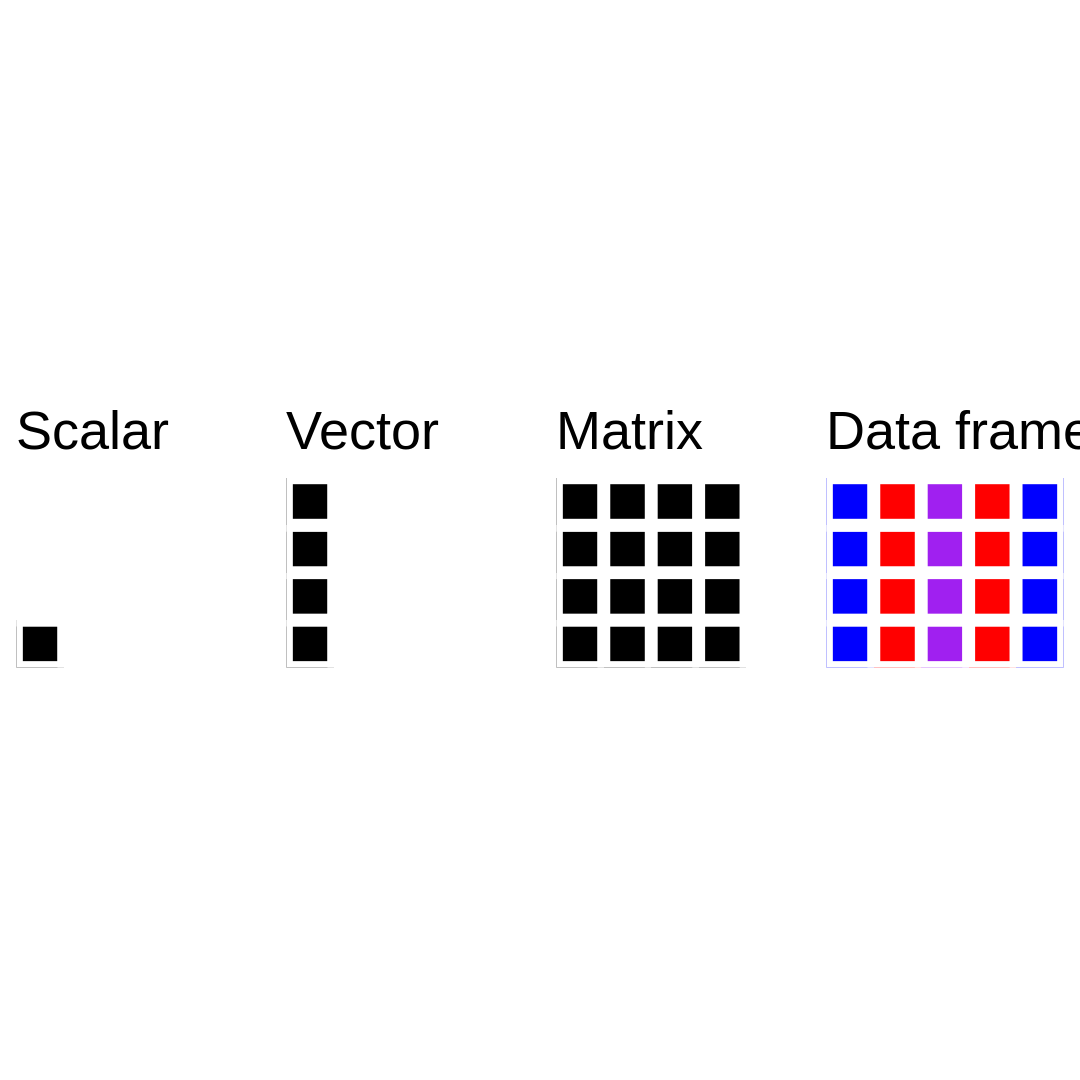
Data structure in R: data frames
Data frames resemble a lot the usual Excel tables that we use in our research!
| site_id | soil_pH | num_sp | fertilised |
|---|---|---|---|
| A1.01 | 5.6 | 17 | yes |
| A1.02 | 7.3 | 23 | yes |
| B1.01 | 4.1 | 15 | no |
| B1.02 | 6.0 | 7 | no |
site_ididentifies the sampling site,soil_pHis the soil pH,num_spis the number of species, andfertilisedidentifies the treatment applied.
One of the ways of representing this table in R, is to create vectors:
site_id <- c("A1.01", "A1.02", "B1.01", "B1.02")soil_pH <- c(5.6, 7.3, 4.1, 6.0)num_sp <- c(17, 23, 15, 7)fertilised <- c("yes", "yes", "no", "no")Data structure in R: data frames
Data frames resemble a lot the usual Excel tables that we use in our research!
| site_id | soil_pH | num_sp | fertilised |
|---|---|---|---|
| A1.01 | 5.6 | 17 | yes |
| A1.02 | 7.3 | 23 | yes |
| B1.01 | 4.1 | 15 | no |
| B1.02 | 6.0 | 7 | no |
site_ididentifies the sampling site,soil_pHis the soil pH,num_spis the number of species, andfertilisedidentifies the treatment applied.
One of the ways of representing this table in R, is to create vectors:
site_id <- c("A1.01", "A1.02", "B1.01", "B1.02")soil_pH <- c(5.6, 7.3, 4.1, 6.0)num_sp <- c(17, 23, 15, 7)fertilised <- c("yes", "yes", "no", "no")We then combine them using data.frame():
soil_fertilisation_data <- data.frame(site_id, soil_pH, num_sp, fertilised)Data structure in R: data frames
Data frames resemble a lot the usual Excel tables that we use in our research!
| site_id | soil_pH | num_sp | fertilised |
|---|---|---|---|
| A1.01 | 5.6 | 17 | yes |
| A1.02 | 7.3 | 23 | yes |
| B1.01 | 4.1 | 15 | no |
| B1.02 | 6.0 | 7 | no |
site_ididentifies the sampling site,soil_pHis the soil pH,num_spis the number of species, andfertilisedidentifies the treatment applied.
soil_fertilisation_data looks like this!
soil_fertilisation_data# site_id soil_pH num_sp fertilised# 1 A1.01 5.6 17 yes# 2 A1.02 7.3 23 yes# 3 B1.01 4.1 15 no# 4 B1.02 6.0 7 noNote how the data frame integrated the name of the objects as column names!
3. Manipulating objects in R
Indexing
Indexing objects in R
We have our information stored as a vector, a matrix, or a data.frame in R.
We will probably be interested in accessing and even subsetting the data based on some criteria.
Let's start with a pretty basic one: the square brackets [ ] and [, ]
We can indicate the position of the values we want to see between the brackets. This is often called indexing or slicing.
Let us see how to index a vector object in R.
Indexing vectors
To index an element within a vector using [], we need to write position number of the element within the brackets [].
For instance, here is our odd_n object:
(odd_n <- seq(1,9, by = 2))# [1] 1 3 5 7 9Indexing vectors
To index an element within a vector using [], we need to write position number of the element within the brackets [].
For instance, here is our odd_n object:
(odd_n <- seq(1,9, by = 2))# [1] 1 3 5 7 9To obtain the value in the second position, we do as follows:
odd_n[2]# [1] 3Indexing vectors
To index an element within a vector using [], we need to write position number of the element within the brackets [].
For instance, here is our odd_n object:
(odd_n <- seq(1,9, by = 2))# [1] 1 3 5 7 9To obtain the value in the second position, we do as follows:
odd_n[2]# [1] 3We can also obtain values for multiple positions within a vector with c():
odd_n[c(2, 4)]# [1] 3 7Indexing vectors
To index an element within a vector using [], we need to write position number of the element within the brackets [].
For instance, here is our odd_n object:
(odd_n <- seq(1,9, by = 2))# [1] 1 3 5 7 9To obtain the value in the second position, we do as follows:
odd_n[2]# [1] 3We can also obtain values for multiple positions within a vector with c():
odd_n[c(2, 4)]# [1] 3 7And, we can remove values pertaining to particular positions from a vector using the minus (-) sign before the position value:
odd_n[-c(1, 2)]# [1] 5 7 9odd_n[-4]# [1] 1 3 5 9Challenge 
Using the vector num_vector and our indexing abilities:
- Extract the 4th value;
- Extract the 1st and 3rd values;
- Extract all values except for the 2nd and the 4th;
- Extract from the 6th to the 10th value.
num_vector <- c(1, 4, 3, 98, 32, -76, -4)Challenge: Solution 
Using the vector num_vector and our indexing abilities:
- Extract the 4th value
num_vector[4]# [1] 98- Extract the 1st and 3rd values
num_vector[c(1, 3)]# [1] 1 3- Extract all values except for the 2nd and the 4th
num_vector[c(-2, -4)]# [1] 1 3 32 -76 -4- Extract from the 6th to the 10th value.
num_vector[6:10]# [1] -76 -4 NA NA NA
What happened there? What is that NA?
Indexing from matrices and data frames
To index a data frame you must specify the position of values within two dimensions: within the row and columns, as in:
data_frame_name[row_number, column_number]In this way, to extract the first row:
my_df[1, ]To extract the third column:
my_df[, 3]And, to extract the element within the second row and the fourth column:
my_df[2, 4]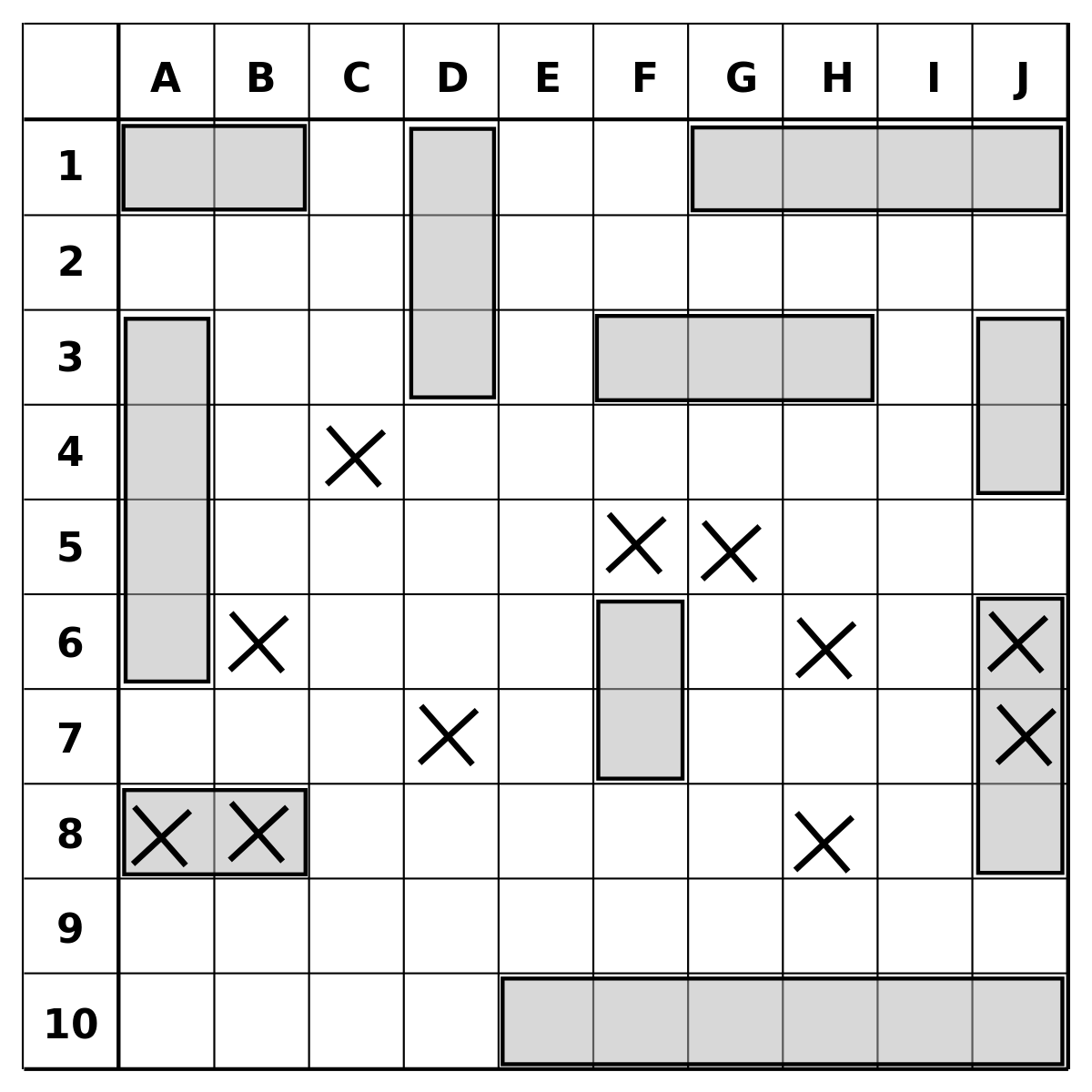
Battleship (game) feelings!
Indexing matrices and data frames by variable names
Remember that our soil_fertilisation_data data frame had column names?
soil_fertilisation_data# site_id soil_pH num_sp fertilised# 1 A1.01 5.6 17 yes# 2 A1.02 7.3 23 yes# 3 B1.01 4.1 15 no# 4 B1.02 6.0 7 no
We can subset columns from it using the column names:
soil_fertilisation_data[ , c("site_id", "soil_pH")]# site_id soil_pH# 1 A1.01 5.6# 2 A1.02 7.3# 3 B1.01 4.1# 4 B1.02 6.0And, also subset columns from it using $:
soil_fertilisation_data$site_id# [1] "A1.01" "A1.02" "B1.01" "B1.02"Note: $ only works for data frames!
What if I want to subset columns based on a condition?
What if I want to subset columns based on a condition?
We can do that! But first, you need to learn about logical operators and logical testing.
Statement testing with logical operators
You might remember about the logical data types, which contains only TRUE and FALSE values.
We can use operators to obtain TRUE or FALSE values for a type of testing. See examples below:
| Operator | Description | Example | Result |
|---|---|---|---|
< and > |
less than or greater than | odd_n > 3 |
FALSE, FALSE, TRUE, TRUE, TRUE |
<= and >= |
less/greater or equal to | odd_n >= 3 |
FALSE, TRUE, TRUE, TRUE, TRUE |
== |
exactly equal to | odd_n == 3 |
FALSE, TRUE, FALSE, FALSE, FALSE |
!= |
not equal to | odd_n != 3 |
TRUE, FALSE, TRUE, TRUE, TRUE |
x | y |
x OR y | odd_n[odd_n >= 5 | odd_n < 3] |
1, 5, 7, 9 |
x & y |
x AND y | odd_n[odd_n >=3 & odd_n < 7] |
3, 5 |
x %in% y |
x match y | odd_n[odd_n %in% c(3,7)] |
3, 7 |
Indexing with logical operators
We can use conditions to select values:
odd_n[odd_n > 4]# [1] 5 7 9Indexing with logical operators
We can use conditions to select values:
odd_n[odd_n > 4]# [1] 5 7 9It is also possible to match a character string.
char_vector <- c("blue", "red", "green")char_vector[char_vector == "blue"]# [1] "blue"Statement testing with logical functions
There are also ways in R that allows us to test conditions!
We can for, instance, test if values within a vector or a matrix are numeric:
char_vector# [1] "blue" "red" "green"is.numeric(char_vector)# [1] FALSEodd_n# [1] 1 3 5 7 9is.numeric(odd_n)# [1] TRUEStatement testing with logical functions
There are also ways in R that allows us to test conditions!
We can for, instance, test if values within a vector or a matrix are numeric:
char_vector# [1] "blue" "red" "green"is.numeric(char_vector)# [1] FALSEodd_n# [1] 1 3 5 7 9is.numeric(odd_n)# [1] TRUEOr, whether they are of the character type:
char_vector# [1] "blue" "red" "green"is.character(char_vector)# [1] TRUEodd_n# [1] 1 3 5 7 9is.character(odd_n)# [1] FALSEStatement testing with logical functions
There are also ways in R that allows us to test conditions!
We can for, instance, test if values within a vector or a matrix are numeric:
char_vector# [1] "blue" "red" "green"is.numeric(char_vector)# [1] FALSEodd_n# [1] 1 3 5 7 9is.numeric(odd_n)# [1] TRUEOr, whether they are of the character type:
char_vector# [1] "blue" "red" "green"is.character(char_vector)# [1] TRUEodd_n# [1] 1 3 5 7 9is.character(odd_n)# [1] FALSEAnd, also, if they are vectors:
char_vector# [1] "blue" "red" "green"is.vector(char_vector)# [1] TRUEChallenge 
Explore the difference between these two lines of code:
char_vector == "blue"char_vector[char_vector == "blue"]Challenge: Solution 
Explore the difference between these 2 lines of code:
char_vector == "blue"# [1] TRUE FALSE FALSEIn this line of code, you test a logical statement. For each entry in the char_vector, R checks whether the entry is equal to blue or not.
char_vector[char_vector == "blue"]# [1] "blue"In this above line, we asked R to extract all values within the char_vector vector that are exactly equal to blue.
Challenge 
Extract the
num_spcolumn fromsoil_fertilisation_dataand multiply its value by the first four values ofnum_vec.After that, write a statement that checks if the values you obtained are greater than 25.
Challenge: Solution 
- Extract the
num_spcolumn fromsoil_fertilisation_dataand multiply its value by the first four values ofnum_vec.
soil_fertilisation_data$num_sp * num_vector[c(1:4)]# [1] 17 92 45 686or
soil_fertilisation_data[, 3] * num_vector[c(1:4)]# [1] 17 92 45 686- After that, write a statement that checks if the values you obtained are greater than 25.
(soil_fertilisation_data$num_sp * num_vector[c(1:4)]) > 25# [1] FALSE TRUE TRUE TRUEOther kinds of data structure: arrays and lists
We focused here mainly on vectors and data frames.
While we will discuss discuss about arrays and lists in other workshops, you can already have an idea what these types of object structure are.
Any wild guesses?
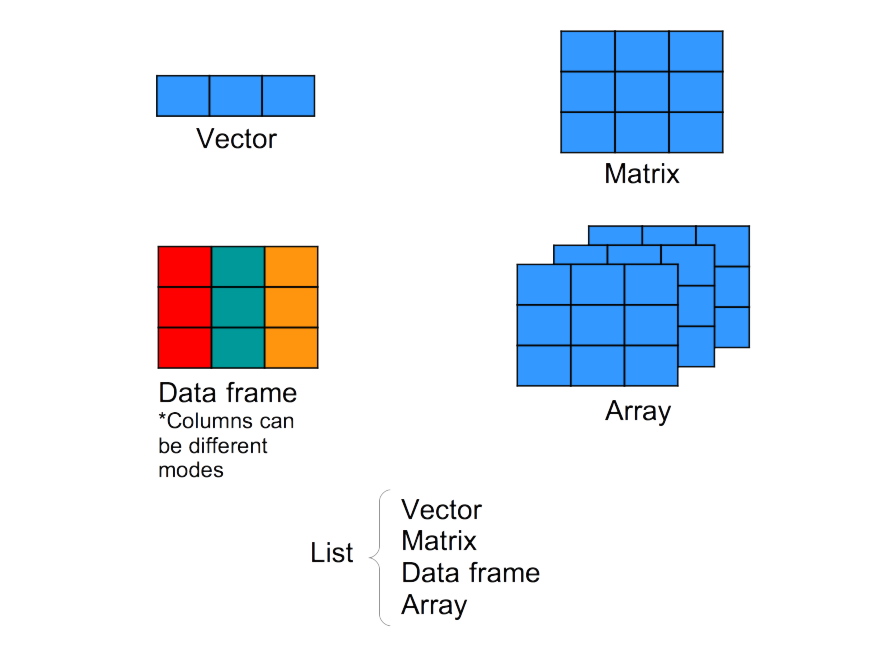
Short review about data structure in R

3. Manipulating objects in R
Built-in functions
Functions
A function is a tool that simplifies our lives!
It allows you to quickly execute operations on objects without having to write every mathematical step.
A function needs entry values called arguments (or parameters).
It then performs (hidden) actions using these arguments and returns an output.
Today, we will look only into
R's built-in functions, but you will learn how to make your own functions during Workshop #5!
Using functions
To use (or to call) a function, the command must be structured properly, following the "grammar rules" of the R language: the syntax.
function_name(argument1 = value, argument2 = value, ..., argument4 = value)Using functions: arguments
Arguments are values and instructions the function needs to run.
Objects storing these values and instructions can be used in functions:
a <- 3b <- 5sum(a, b)# [1] 8mean(soil_fertilisation_data$num_sp)# [1] 15.5Challenge 
- Create a vector
athat contains all the numbers from 1 to 5. - Create an object
bthat has a value of 2. - Add
aandbtogether using the basic+operator and save the result in an object calledresult_add. - Add
aandbtogether using thesumfunction and save the result in an object calledresult_sum. - Are
result_addandresult_sumdifferent? - Add
5toresult_sumusing thesum()function.
Challenge: Solution 
- Are
result_addandresult_sumdifferent?
a <- c(1:5)b <- 2result_add <- a + bresult_sum <- sum(a, b)- Add
5toresult_sumusing thesum()function.
result_add# [1] 3 4 5 6 7result_sum# [1] 17sum(result_sum, 5)# [1] 22Challenge: Solution 
- Are
result_addandresult_sumdifferent?
a <- c(1:5)b <- 2result_add <- a + bresult_sum <- sum(a, b)- Add
5toresult_sumusing thesum()function.
result_add# [1] 3 4 5 6 7result_sum# [1] 17sum(result_sum, 5)# [1] 22The operation + on the vector a adds 2 to each element. The result is a vector.
The function sum() concatenates all the values provided and then sum them. It is the same as doing 1 + 2 + 3 + 4 + 5 + 2.
Arguments
Each argument has a name, which may be used during a function call.
For instance, the first arguments of the matrix() function are:
matrix(data, nrow, ncol)We can create a matrix using:
matrix(data = 1:12, nrow = 3, ncol = 4)# [,1] [,2] [,3] [,4]# [1,] 1 4 7 10# [2,] 2 5 8 11# [3,] 3 6 9 12We can also execute a function when omitting argument names, however the order of the values will matter:
matrix(data = 1:12, 3, 4)# [,1] [,2] [,3] [,4]# [1,] 1 4 7 10# [2,] 2 5 8 11# [3,] 3 6 9 12matrix(1:12, 4, 3)# [,1] [,2] [,3]# [1,] 1 5 9# [2,] 2 6 10# [3,] 3 7 11# [4,] 4 8 12Challenge 
plot is a function that draws a graph of y as a function of x. It requires two arguments names x and y. What are the differences between the following lines?
a <- 1:100b <- a^2plot(a, b, type = "l")plot(b, a, type = "l")plot(x = a, y = b, type = "l")plot(y = b, x = a, type = "l")The argument type of the function plot let you choose the type of graph you want. Try it without this argument.
Challenge: Solution 
plot is a function that draws a graph of y as a function of x. It requires two arguments names x and y. What are the differences between the following lines?
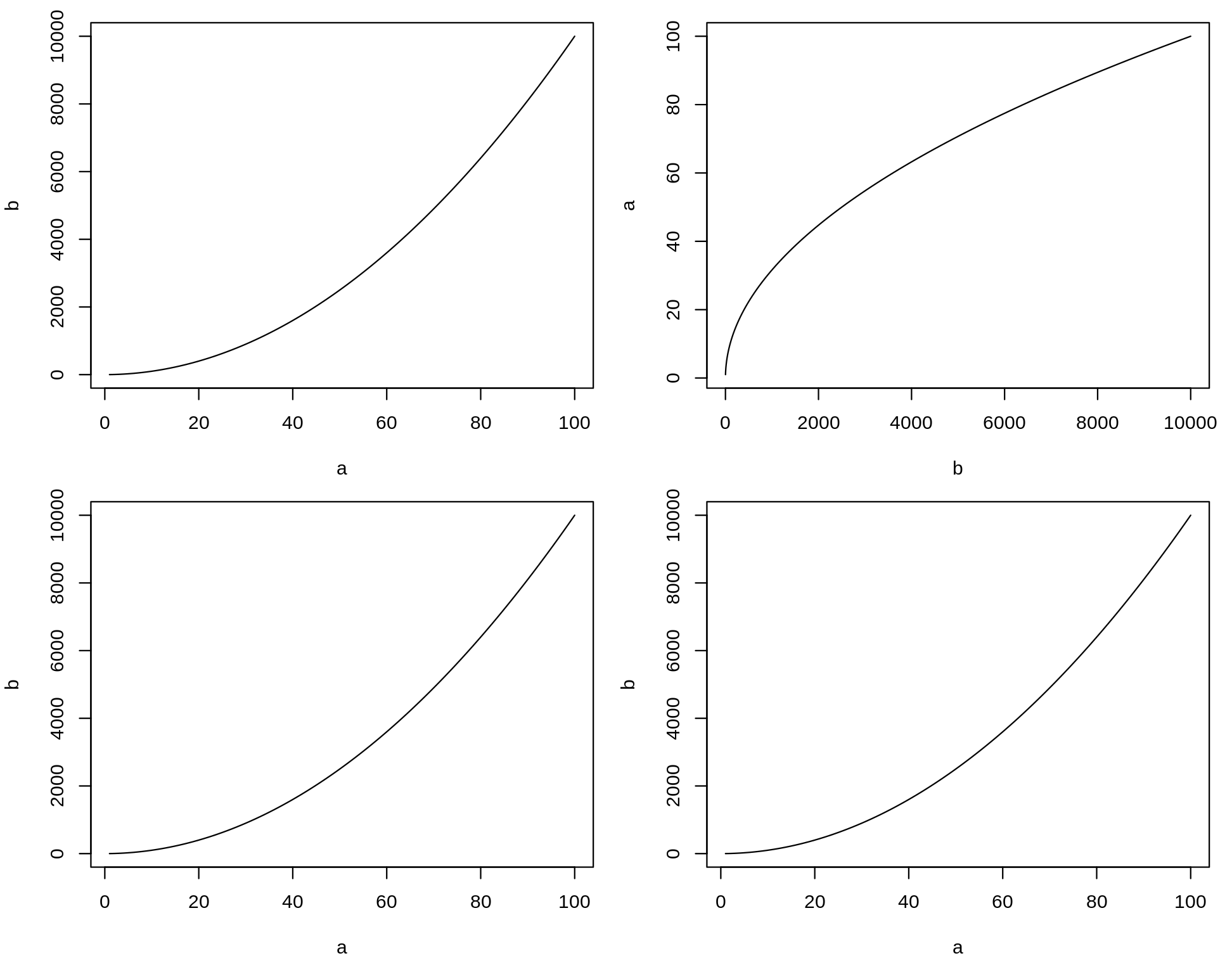
Challenge: Solution
plot(a, b, type = "l")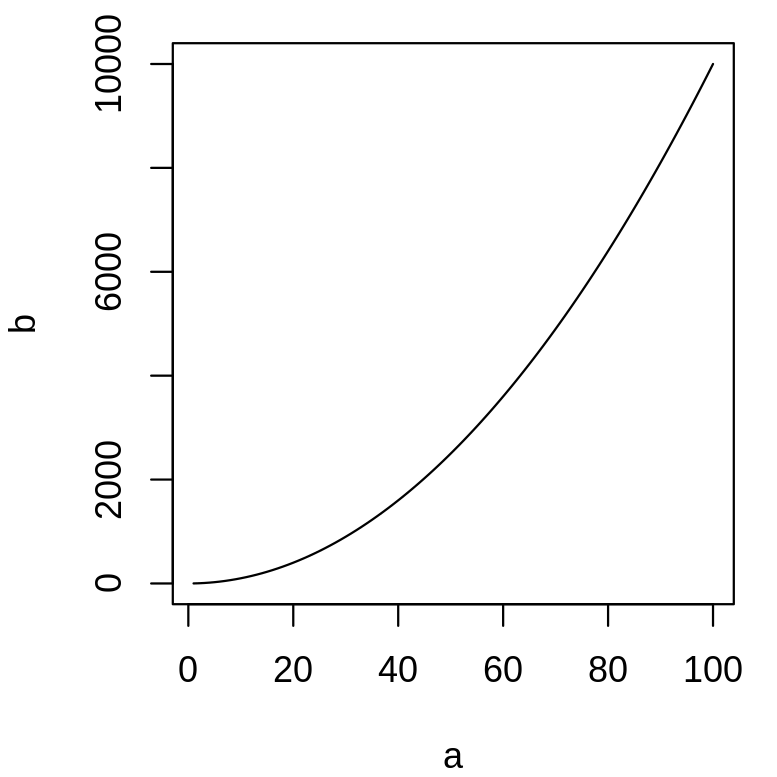
plot(b, a, type = "l")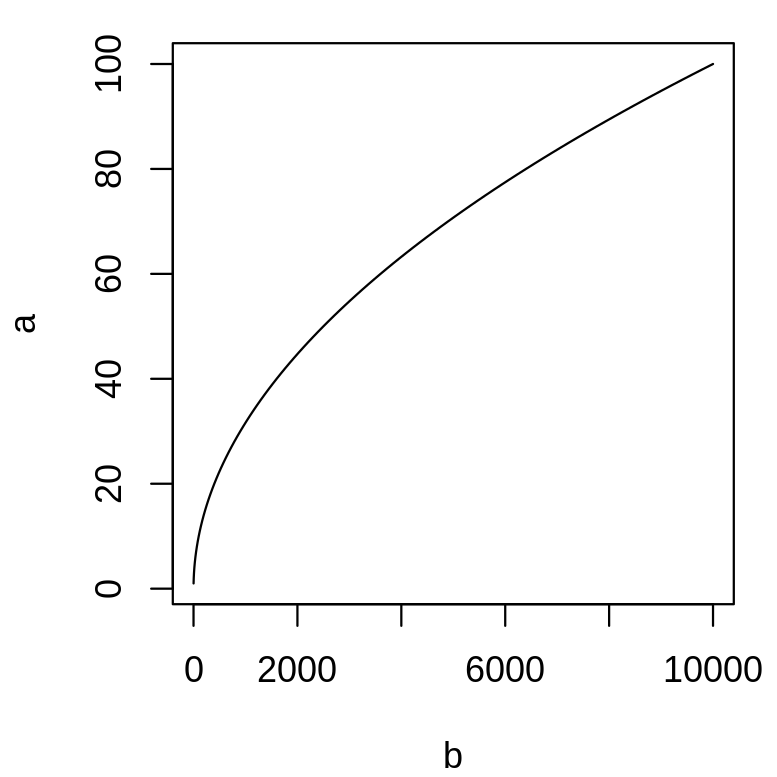
The shape of the plot changes, as we did not provide the argument's names, the order is important.
Challenge: Solution
plot(x = a, y = b, type = "l")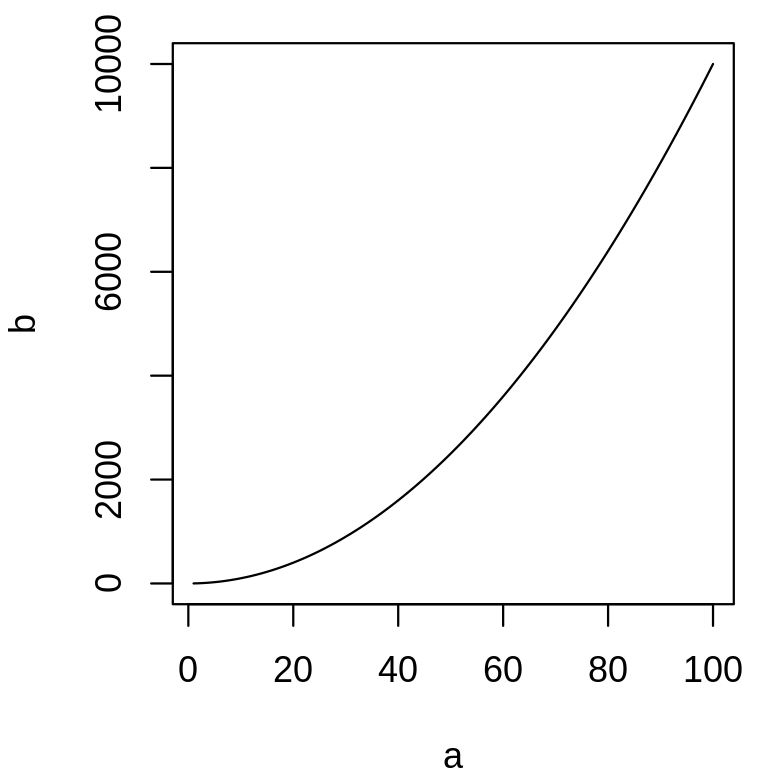
plot(y = b, x = a, type = "l")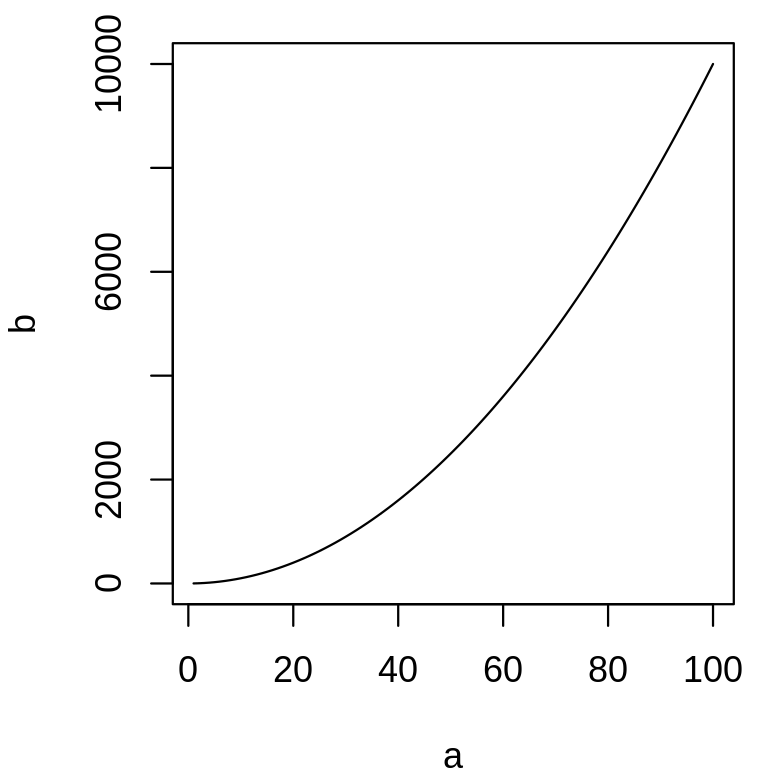
Same as plot(a, b, type = "l"). The argument names are provided, the order is not important.
4. Installing and using R packages
R Packages
Packages group functions and/or datasets that share a similar theme, e.g. statistics, spatial analysis, plotting.
Anyone can develop packages and make them available to others.
Many packages available through the Comprehensive R Archive Network (CRAN) and now many more on GitHub.
Guess how many packages are available (not only within the CRAN)?
Installing R packages
To install packages in your Computer, use the function install.packages().
install.packages("package_name")Installing a package is essential to use it, but there is one more step: loading it.
You can load a package into your workspace using the library() function.
library(package_name)Installing your first R package: ggplot2
Let us install a popular visualization package called ggplot.
install.packages("ggplot2")Installing package into '/home/labo/R/x86_64-redhat-linux-gnu-library/3.3'(as 'lib' is unspecified)Now we will use the function qplot from the package
qplot(1:10, 1:10)Installing your first R package: ggplot2
Let us install a popular visualization package called ggplot.
install.packages("ggplot2")Installing package into '/home/labo/R/x86_64-redhat-linux-gnu-library/3.3'(as 'lib' is unspecified)Now we will use the function qplot from the package
qplot(1:10, 1:10)Did you get this error?
## Error: could not find function "qplot"Loading your first R package: ggplot2
We need to gain access the functions that are available within the installed package. To do this, load ggplot2 using the library() function.
library(ggplot2)Now we can draw the graph
qplot(1:10, 1:10)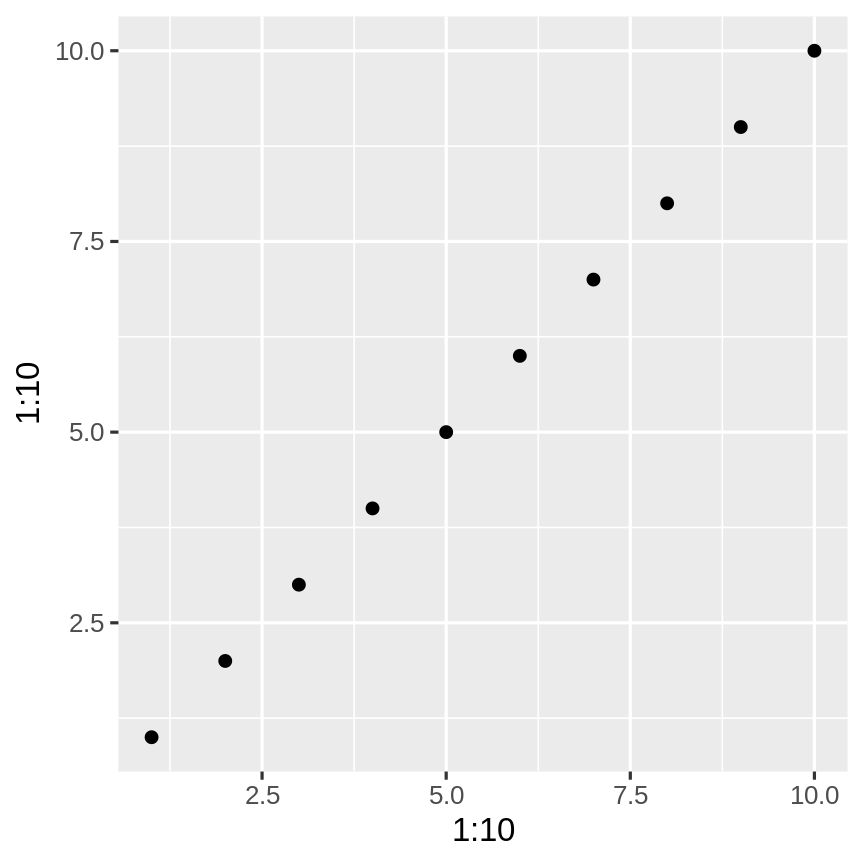
The ggplot2 package will be covered in Workshop #3: Introduction to ggplot2.
Finding functions within packages
WOW! R is so great! So many functions to do what I want!
But... how do I find them?
Finding functions within packages
WOW! R is so great! So many functions to do what I want!
But... how do I find them?
To find a function that does something specific in your installed packages, you can use ?? followed by a search term.
Let us say we want to create a sequence of odd numbers between 0 and 10 as we did earlier. We can search in our packages all the functions with the word "sequence" in them:
??sequenceSearch results
Search results
Search results
Getting help with functions
OK! So let us use the seq function!
But wait... how does it work? What arguments does it need?
To find information about a function in particular, use ?
?seqHelp pages
Description
function_name {package_name}Description: a short description of what the function does.

Usage
- How to call the function
- If
name = valueis present, a default value is provided if the argument is missing. The argument becomes optional. - Other related functions described in this help page

Arguments
- Description of all the arguments and what they are used for

Details
- A detailed description of how the functions work and their characteristics

Value, See Also, and Examples
- A description of the return value

Value, See Also, and Examples
- A description of the return value

- Other related functions that can be useful

Value, See Also, and Examples
- A description of the return value

- Other related functions that can be useful

- Reproducible examples

Challenge 
Create a sequence of even numbers from 0 to 10 using the
seqfunction.Create an unsorted vector of your favourite numbers, then sort your vector in reverse order.
Challenge: Solutions 
- Create a sequence of even numbers from 0 to 10 using the
seqfunction.
seq(from = 0, to = 10, by = 2)# [1] 0 2 4 6 8 10seq(0, 10, 2)# [1] 0 2 4 6 8 10- Create an unsorted vector of your favorite numbers, then sort your vector in reverse order.
numbers <- c(2, 4, 22, 6, 26)sort(numbers, decreasing = TRUE)# [1] 26 22 6 4 2Other ways to get help
Usually, your best source of information will be your favorite search engine!
Here are some tips on how to use them efficiently:
- Search in English;
- Use the keyword
Rat the beginning of your search; - Define precisely what you are looking for;
- Learn to read discussion forums, such as StackOverflow. Chances are other people already had your problem and asked about it!
- Do not hesitate to search again using different keywords!
Challenge 
Find the appropriate functions to perform the following operations:
- Square root
- Calculate the mean of numbers
- Combine two data frames by columns
- List available objects in your workspace
Challenge: Solutions 
Find the appropriate functions to perform the following operations:
- Square root
sqrt()
- Calculate the mean of numbers
mean()
- Combine two data frames by columns
cbind()
- List available objects in your workspace
ls()
Additional resources
Cheat 4ever

Lots of cheat sheets are available online.
Open it directly from RStudio: Help → Cheatsheets
Cheatsheet 4ever
Some useful R books






Thank you for attending!